1. Introduction
The ability to accurately date archaeological sites and their material is essential to the study of human evolution over the Pleistocene. Caves are common archaeological sites within the Palaeolithic (e.g. Barham and Mitchell, 2008; Dennell, 2008; French, 2021) and can act as excellent fossil repositories, especially in areas of the African continent such as the South-Central region (Figure 1), where fossil preservation is generally poor (Bishop et al., 2016). This region’s current ecology, reflecting its topography, is diverse, with tropical deciduous woodlands across the high central plateau grading to dryland savannah on the margins of the Kalahari basin. Large wetlands occur in northern Zambia and in neighbouring northern Botswana today, and evidence has been found for a vast palaeo-wetland during the Pleistocene encompassing the Okavango delta, Makgadikgadi basin, and the Zambezi and Kafue rivers (Chan et al., 2019; Burrough et al., 2022). This wetland would have provided both a migratory corridor between South and East Africa and an ideal environment for mammalian (including hominin) habitation. The region has preserved important Mid-Pleistocene fossils, most notably the Kabwe cranium (Homo heidelbergensis (rhodesiensis)) (Grün et al., 2020), evidence for the transition from the Early to Middle Stone Age (Duller et al., 2015), the preservation of evidence for early pigment use (Barham, 2002) and woodworking (Barham et al., 2023).
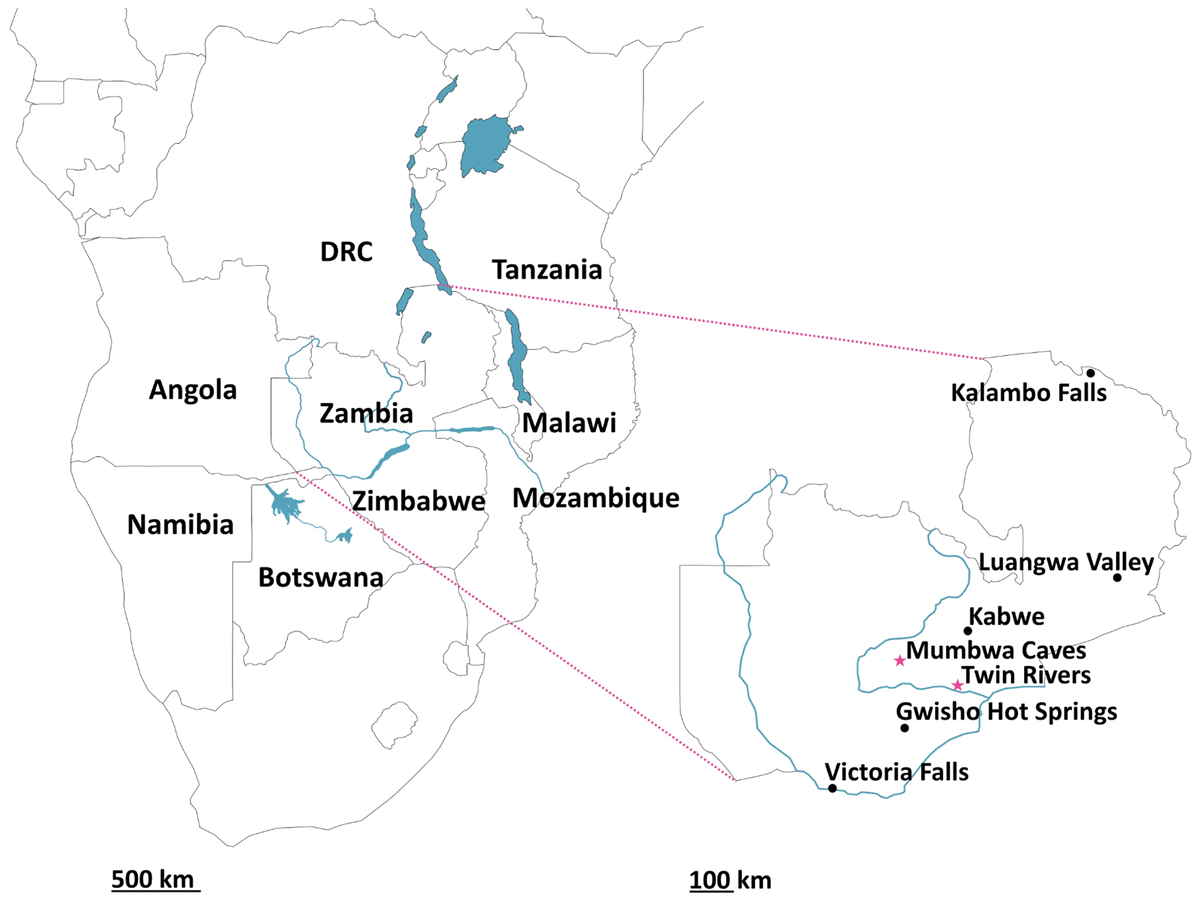
Figure 1
Left: Map of southern Africa highlighting the South-Central African region with major current water bodies; Right: Map of Zambia highlighting key archaeological sites. Those included in this study (Twin Rivers and Mumbwa Caves) are highlighted with pink stars.
Many archaeological and palaeoenvironmental sites have poorly constrained chronologies, and cave sites are well known for their complex depositional histories, making dating and accurate site interpretation especially challenging (e.g. Brain, 1983; Reynolds and Kibii, 2011; Val et al., 2015; Marin-Monfort et al., 2022). Direct dating of fossils (as opposed to dating associated sediments and/or flowstones) can enable identification of stratigraphic disruption and place the archaeologically/palaeontologically relevant material within a more detailed stratigraphic context. One such technique, amino acid geochronology (AAG), has the potential to provide direct, relative, dating on fossil biominerals (e.g. Wehmiller, 1977; Bowen et al., 1989; Brooks et al., 1990; Penkman et al., 2007; Hearty and Kaufman, 2009; Torres et al., 2014; Koppel et al., 2016). AAG exploits the time-dependent degradation of proteins (such as amino acid racemisation; AAR) trapped in fossil biominerals (Abelson, 1955; Hare and Mitterer, 1967). As protein degradation is temperature-dependent (Wehmiller and Miller, 2000) AAGs are built regionally for each specific biomineral, and to date only one AAG study, using gastropod shell from sediments associated with Palaeolake Kafue, has been published within the South-Central African region (Baldreki et al., 2024).
One approach which has been shown to improve the reliability of AAG data in some biominerals is intra-crystalline protein degradation (IcPD), which targets the analysis of protein trapped within the fossil’s mineral matrix. This fraction of protein, isolated with the use of a strong chemical oxidant, has been shown to effectively operate as a closed system in many CaCO3 biominerals (e.g. gastropods (Sykes et al., 1995; Penkman et al., 2008; Demarchi et al., 2013a), ostrich eggshell (Crisp et al., 2013a), coral (Hendy et al., 2012) and foraminifera (Wheeler et al., 2021)). Where the intra-crystalline fraction is shown to operate as a closed system, leaching of endogenous protein, contamination by exogenous protein and most additional environmental impacts on protein degradation (e.g. pH (Hare and Mitterer, 1969)) are minimised (Towe, 1980). In some biominerals, targeting the intra-crystalline fraction has been shown to improve the accuracy and precision of the data, thereby increasing the reliability of the geochronologies obtained (e.g. Brooks et al., 1990; Penkman et al., 2013; Wheeler et al., 2021). Oxidative treatment is not always suitable or necessary (e.g. Orem and Kaufman, 2011; Torres et al., 2014; Demarchi et al., 2015; Ortiz et al., 2017), and stringent data screening approaches have also been successfully employed for many fossil biominerals (e.g. Brooks et al., 1990; Kaufman, 2006; Kosnik and Kaufman, 2008; Ortiz et al., 2013; 2018; Wheeler, 2022).
Recent methodological advances to the IcPD protocol for the analysis of tooth enamel (CaPO4) has confirmed that this biomineral also has an intra-crystalline protein fraction which effectively behaves as a closed system (Dickinson et al., 2019). This has opened an additional avenue of direct dating for fossil teeth, which are commonly found at archaeological cave sites. The enamel proteome is composed of a small number of proteins, whose primary sequence has been shown to vary between taxa (and therefore clarify phylogenetic relationships e.g. Cappellini et al., 2019; Welker et al., 2019). As protein degradation, including racemisation, is influenced by the sequence of amino acids in a peptide chain (Mitterer and Kriausakul, 1984), any taxonomic effect in tooth enamel IcPD rates, need to be accounted for in any arising amino acid geochronology (Cappellini et al., 2019; Dickinson et al., 2024).
Here we explore the capacity of tooth enamel to build taxon-specific aminostratigraphies for the South-Central African region using IcPD analysis. We tested fossil teeth/fragments of four commonly occurring taxonomic groups (rhinocerotid, equid, suid, bovid) excavated from Twin Rivers and Mumbwa Caves archaeological sites in Zambia (Figure 1). We investigate whether the intra-crystalline fraction of protein behaves as a closed system in these taxa and examine age-related signals of protein degradation. Through these investigations, we demonstrate the use of this AAG approach to examine depositional histories and taphonomies, and inter-site relationships.
2. Materials & Methods
2.1. Materials
The 80 fossil teeth/fragments included in this study are from two archaeological cave sites in Zambia (Twin Rivers and Mumbwa Caves; Figure 1, Table 1, SI Data) and represent commonly found fauna in the region. Both sites provide important regional climate and hominin behavioural records: Mumbwa Caves for the Middle to Late Pleistocene (Burrough et al., 2019) and Twin Rivers for the Middle Pleistocene (Barham, 2000). Both sites were excavated in the 1990s with faunal assemblages reported and discussed in Barham (2000), Klein and Cruz-Uribe (2000), and Bishop and Reynolds (2000).
Table 1
Tooth enamel samples analysed in this study; numbers in brackets indicate the number of individual teeth/fragments sampled. For more detailed sample information please see the supplementary data.
| SITE NAME | EXCAVATION | TAXON | |
|---|---|---|---|
| CAVITY | LEVEL | ||
| Twin Rivers | E5/F5 | 1 | Rhinocerotid (3), Suid (1), Equid (1), Bovid (1) |
| 2 | Bovid (1) | ||
| E4/F4 | 1 | Suid (3), Equid (6) | |
| 3 | Rhinocerotid (3), Equid (1), Bovid (1) | ||
| E5/E6 | 1 | Rhinocerotid (3), Equid (1) | |
| 2 | Rhinocerotid (2), Equid (4) | ||
| 3 | Rhinocerotid (5), Suid (2), Bovid (4) | ||
| 4 | Rhinocerotid (1), Suid (2), Equid (3) | ||
| 5 | Equid (1), Bovid (1) | ||
| G5 | N/A | Bovid (3) | |
| Mumbwa Caves | N/A | N/A | Suid (10), Equid (4), Bovid (13) |
2.1.1. Mumbwa Caves
Mumbwa Caves in Zambia has had a number of dating techniques applied to its sediments and materials (Barham and Debenham, 2000) including thermoluminescence (TL) on burnt quartz and calcite, radiocarbon on charcoal, optically stimulated luminescence (OSL) on sediment and electron spin resonance (ESR) on enamel, and the stratigraphy is relatively well constrained to a maximum age of >172 ka (Barham, 2000).
Amino acid geochronology was considered during the 1993 excavation, but the only applicable biomineral at the time was ostrich eggshell, which was not present at the site (Brooks et al., 1990; Barham and Debenham, 2000). The enamel IcPD protocol published in 2019 (Dickinson et al., 2019) therefore opened another avenue for dating and depositional history interpretation. Unfortunately, the teeth/fragments used for the previous enamel ESR analysis were not available, and thus IcPD analysis could not be undertaken on these specimens for direct comparison between the two dating techniques. Additionally, the stratigraphic context of each specimen that was available for analysis was unrecoverable, and the loss of contextual information (including taxonomy) meant that individual specimens could not be linked to the published identifications. One intention of this study was therefore to increase the scientific value of these materials through re-identification of their taxonomy and explore their potential to provide useful age information through IcPD analysis.
2.1.2. Twin Rivers
The tooth enamel samples analysed in this study from Twin Rivers were excavated from A Block (Figure 2) in 1999. A Block originated as an irregularly shaped phreatic passage and the sediments are thought to have been deposited in periodic, gentle slurry flows. Over time much of the deposits became cemented into breccia with lenses of speleothem separating the breccia into units. The cave roof collapsed at an unknown time, leaving the deposit exposed to weathering. Dynamite was used in the 1950s to excavate much of the deposit (Clark and Brown 2001; blast scree area shown in Figure 2), though none of the material included in this study comes from this excavation. The 1999 excavations focused on shallow (<60 cm) and narrow (<30 cm) pockets of loose sandy sediment between the breccia and the cave walls. The pockets contained sharp stone tools, ochre pieces, and faunal fossils (Barham, 2000). As a hilltop site, the presence of rhinocerotid and giraffid (among others) within the fossil assemblage is thought to have accumulated from predation and/or scavenging activity, including the possibility of (but not limited to) hominin activity. The excavation was named using a hybrid reference system (e.g. E5/E6; Figure 2), with each pocket/cavity discrete from one another. The excavation within each discrete cavity was conducted in horizontal levels in the absence of any visible stratigraphy. There is therefore no correlation between excavation levels of different cavities.
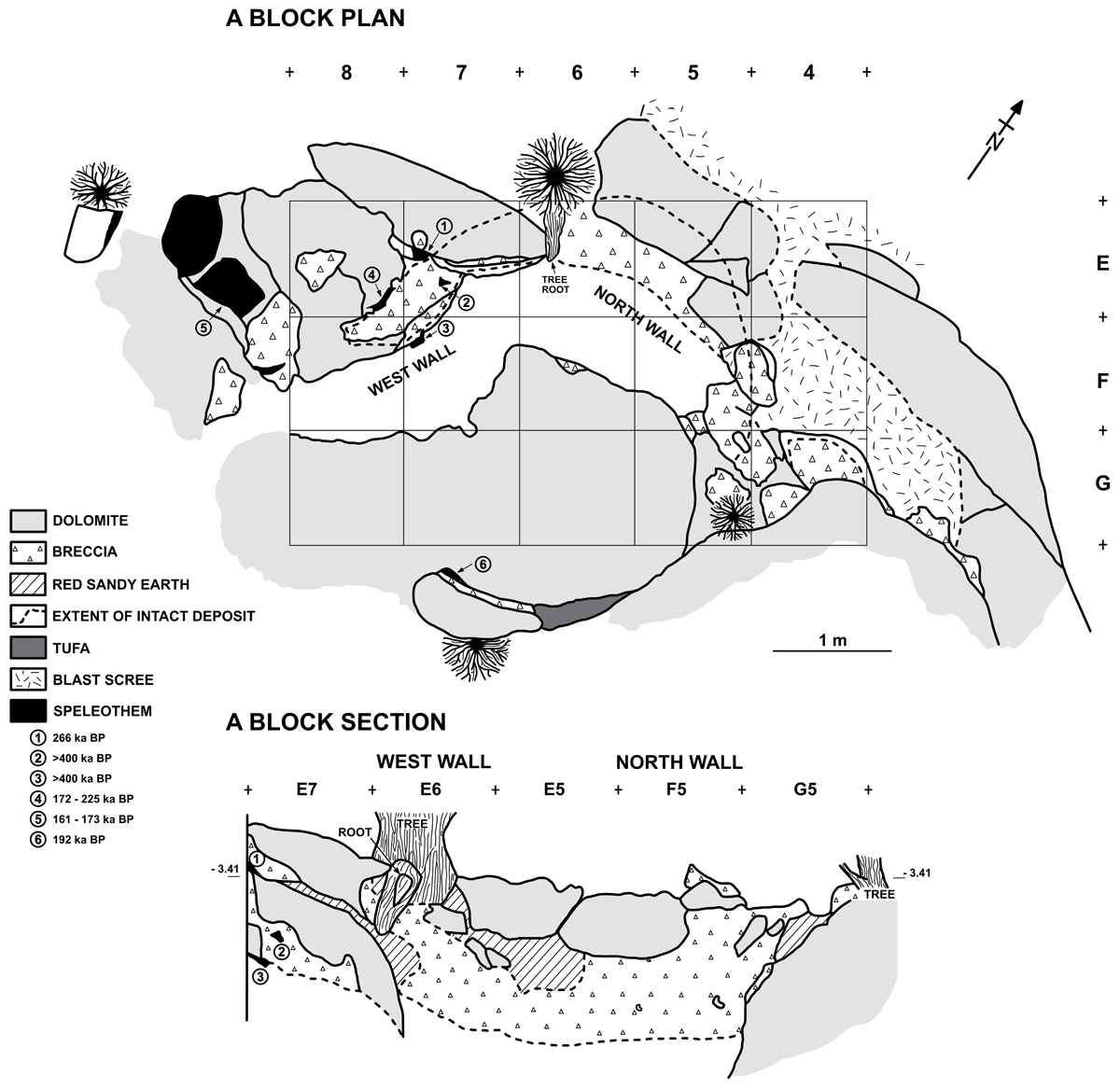
Figure 2
Plan and section (west and north walls) of Twin Rivers A Block excavated in 1999. Previously dated speleothem samples are shown with associated uranium-thorium dates (sample 1 = 266 ka BP, sample 2 = >400 ka BP, sample 3 = >400 ka BP, sample 4 = 172–225 ka BP, sample 5 = 161–173 ka BP, sample 6 = 192 ka BP). All tooth enamel samples analysed for AAG were excavated from red sandy earth sediments which filled multiple discrete cavities between the cave walls and the rim of the intact breccia. Due to the irregular shape of the cave passage and deposits within, not all excavation cavities and no excavation levels are visible in this section.
Previous speleothem U-series dating at the site placed the collapsed cave passage sequence to ~170- >400 ka (Barham, 2000). The nature of the site meant that, with one exception (266 ka), the flowstones were not directly related to the excavated levels. Whilst their associations were helpful in providing minimum and maximum ages, and ages concordant with increasing depth, they were difficult to directly relate to the archaeological material. Additionally, due to the upper age limitation of the U-series dating undertaken here (>400 ka), the base of the sequence has a minimum age of 400 ka but could represent a much older formation. It was therefore hoped that enamel IcPD could be used as a direct dating technique to improve the understanding of the ages of the faunal material from this site.
2.1.3. Taxonomic re-identification
Both the Mumbwa Caves and Twin Rivers teeth/fragments required taxonomic re-identification due to the loss of contextual information. These were visually evaluated by one of us (SCR) who had previously analysed the original faunal material from Twin Rivers in 2000 (Bishop and Reynolds, 2000). Additional published species lists, such as Clark and Brown (2001) were also consulted. The very fragmentary material was assigned to genus level only, and specimens were only assigned to species level if the dental morphology was complete enough to permit this (SI Data). For all subsequent sections, data is discussed in terms of the four family groups commonly identified amongst the Twin Rivers and Mumbwa Caves faunal fossil assemblages (rhinocerotid, equid, suid, bovid). These taxa were selected as they had not yet been characterised for enamel amino acid geochronology but form a common component of Pleistocene faunal assemblages in Africa (and elsewhere) and therefore potentially have a wide utility for dating.
2.2. Enamel IcPD methodology
All tooth enamel samples were prepared and analysed following the methods of Dickinson et al. (2019).
Approximately 40 mg of enamel (as chips, from a single location) was removed from each tooth/fragment (Table 1) using abrasive rotary drill bits on a handheld rotary tool (Dremel drill), before being washed with water (ultrapure, 18.2 MΩ cm–1) and left to air dry. Each enamel sample was powdered with an agate pestle and mortar.
Bleach (12% NaOCl (analytical grade), 50 µL/mg) was added to each accurately weighed powdered sample (in separate 2 mL plastic microcentrifuge tubes) and left on a rotor (constant mild agitation) for 72 hours. The bleach was removed by pipette and each sample was washed five times with water (ultrapure, 18.2 MΩ cm–1), before a final wash with methanol (HPLC-grade) and the powdered sample left to air dry.
Ca. 5 mg subsample replicates (accurate masses noted) of each powdered and bleached sample were weighed out for the analysis of both the free amino acid fraction (FAA) and the total hydrolysable amino acid fraction (THAA). THAA subsample replicates were demineralised in 7 M HCl (20 µL/mg), the vials flushed with N2 and heated in an oven at 110°C for 24 hours, prior to drying by centrifugal evaporation. The THAA subsample replicates were then redissolved in 1 M HCl (20 µL/mg) and the FAA subsample replicates demineralised in 1 M HCl (25 µL/mg), prior to the addition of 1 M KOH (28 µL/mg) to all subsamples, which formed a monophasic translucent gel with a viscous consistency. All subsamples were centrifuged at 13,000 rpm for 5 minutes whereupon a biphasic solution formed (supernatant above a cloudy gel). The supernatant was removed and dried by centrifugal evaporation.
The subsamples were rehydrated in the minimum possible volume (starting at 10 µL/mg, with 10 µL aliquot additions to achieve dissolution where necessary) of a solution of internal standard (L-homo-arginine (0.01 M), hydrochloric acid (0.01 M) and sodium azide (1.5 mM)). Separation of the chiral isomers of the amino acids (Figure 3) was carried out by fluorescence detection RP-HPLC (Agilent 1100) using a modified Kaufman and Manley (1998) method (Penkman, 2005). As experimental subsample replicates were prepared, no analytical replicates were undertaken. Analytical replicates have been shown to account for only a small portion of the total variability, hence the use of subsample experimental replicates also encompasses analytical variability (Powell et al., 2013).
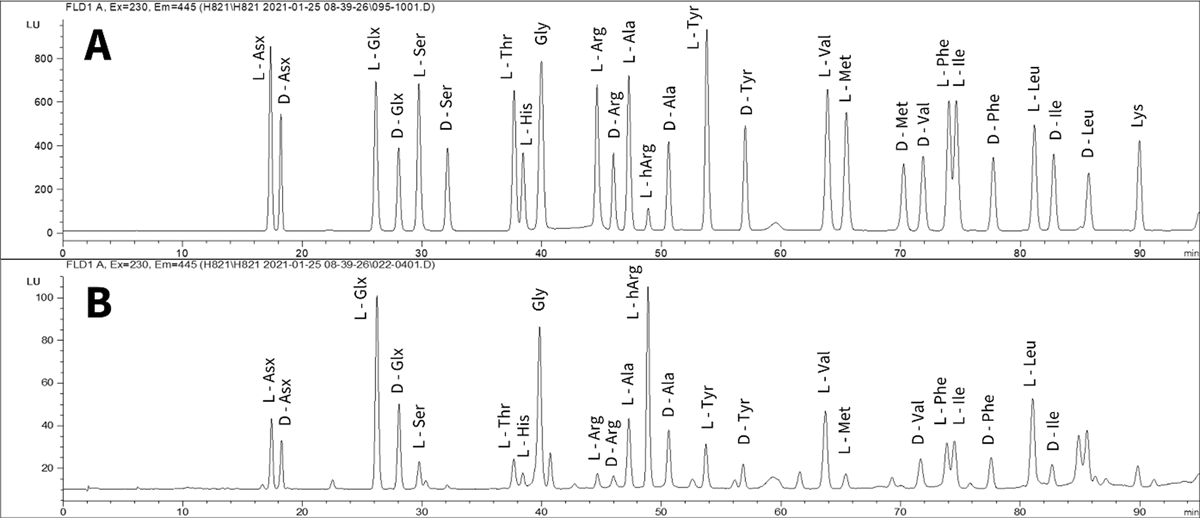
Figure 3
Example chromatograms of A – 0.5 standard, B – rhinocerotid tooth enamel THAA sample.
Total amino acid concentration and relative composition calculations were carried out from the enantiomeric chromatographic peaks of Asx, Glx, Ser, Thr, His, Gly, Arg, Ala, Tyr, Val, Met, Phe, Ile and Leu. In the case of Thr and His only the L enantiomer is resolved by this chromatographic method, whilst Gly contains no stereogenic centre and therefore does not have L/D enantiomers (Figure 3). During the laboratory preparation procedures, irreversible deamidation of Asn to Asp and Gln to Glu is induced. It is therefore not possible to distinguish between these chromatographically and are therefore reported as Asx and Glx (Hill, 1965). Sufficient concentration and chromatographic resolution was achieved for the calculation of racemisation and percentage free values for Asx, Glx, Ser, Ala, Val, Phe within the tooth enamel (SI Data).
3. Results & Discussion
3.1. Assessment of closed-system behaviour
The closed-system behaviour of the intra-crystalline fraction of protein from the 80 enamel samples was assessed by looking at key aspects of IcPD data (Penkman et al., 2013): the relationship between the free and total hydrolysable fractions of amino acids (section 3.1.1); the relative racemisation rates (section 3.1.2); peptide bond hydrolysis (section 3.1.3); serine decomposition (section 3.1.4) and concentration and relative composition (section 3.1.5).
3.1.1. Amino acid fraction D/L covariance
When protein degradation occurs within a closed system, a strong positive correlation should be observed in the extent of racemisation between the free and total hydrolysable amino acid fractions (Preece and Penkman, 2005; Penkman et al., 2007). This was observed for the majority of samples (72) in all four taxonomic groups (rhinocerotid, equid, suid, bovid); two amino acids, Asx and Glx, are provided as examples in Figure 4 (see all additional amino acids in SI Figs. 1–4). This indicates maintenance of closed-system behaviour in these 72 samples over their depositional history. However, eight samples (10%) deviated from the general trend, with large subsample replicate variability observed for the majority of amino acids in these samples (TR12- Figure 4A; TE2, TE6, TE10, TE12- Figure 4B; TS6- Figure 4C; MB1, MB2- Figure 4D). For seven of these samples, in each case one subsample replicate fell within, and one well outside of the observed correlation range (Figure 4). Open-system behaviour from mineral diagenesis can lead to larger variability in results (even within the same sample), and/or off-trend racemisation values (Penkman et al., 2007; Dickinson et al., 2024) and may therefore be an explanation for this data. For one sample (MB1- Figure 4D), unexpectedly high racemisation was observed in only Glx FAA, although replicate precision was high. This sample is discussed further in sections 3.1.2 and 3.1.5. In either case, these eight samples highlight the importance of undertaking preparative replicate analysis to help identify outliers.
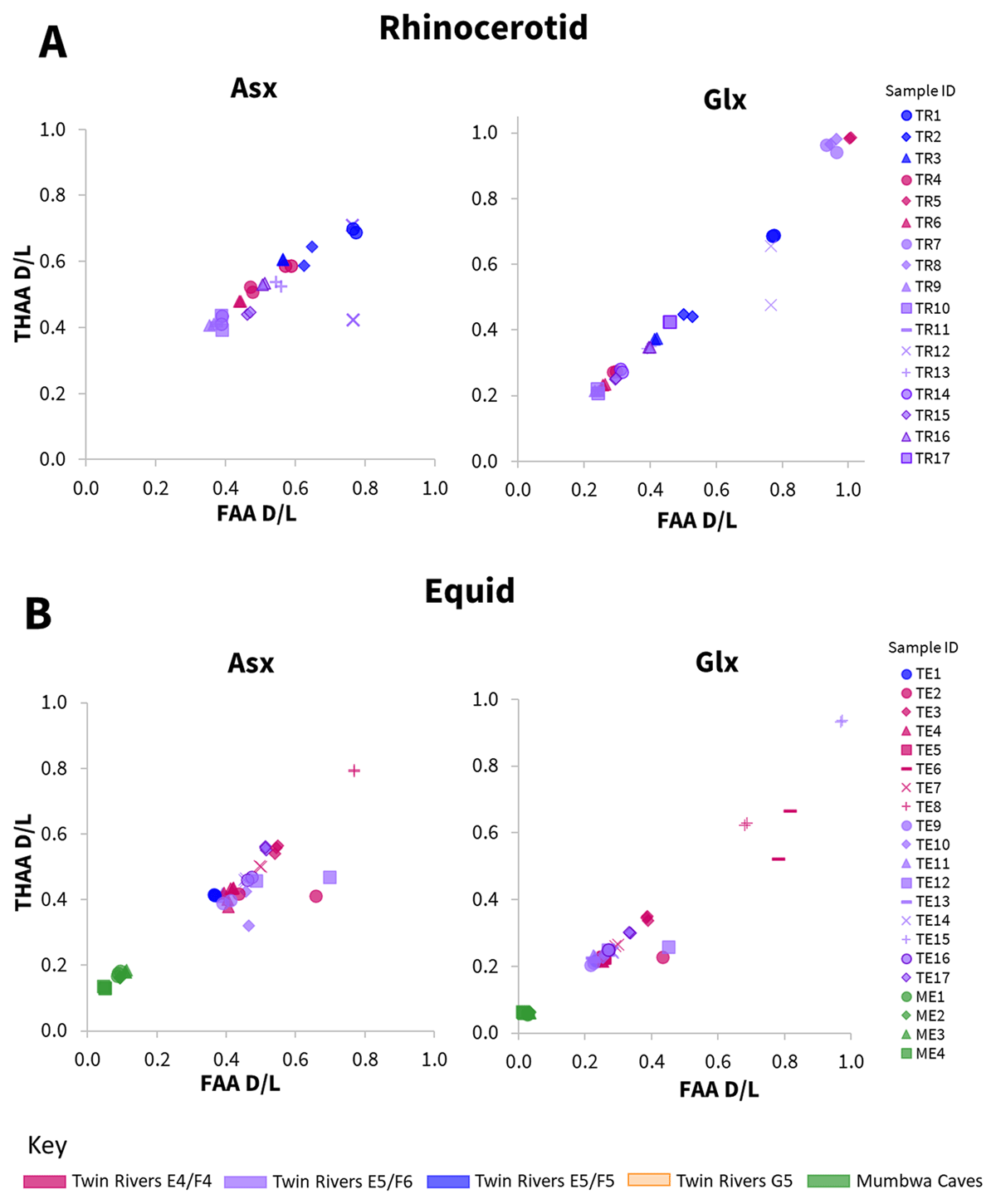
Figure 4a
a Relationship between racemisation values (D/L) in free (FAA) vs total hydrolysable (THAA) amino acids, for subsample replicates, in two taxonomic groups of enamel samples (A – rhinocerotid; B – equid), for two example amino acids (Asx, Glx). Mumbwa Caves samples were all excavated from one deposit (Barham, 2000). Twin Rivers samples were excavated from four areas within the A Block cave passage (Barham, 2000).

Figure 4b
Relationship between racemisation values (D/L) in free (FAA) vs total hydrolysable (THAA) amino acids, for subsample replicates, in two taxonomic groups of enamel samples (C – suid; D – bovid), for two example amino acids (Asx, Glx). Mumbwa Caves samples were all excavated from one deposit (Barham, 2000). Twin Rivers samples were excavated from four areas within the A Block cave passage (Barham, 2000).
3.1.2. Relative rates of racemisation
An additional avenue that can corroborate maintenance of closed-system behaviour is the consistent relative order of amino acid racemisation. Where samples display inconsistencies in the extent of racemisation between amino acids with different relative rates, contamination and/or leaching could be the cause. Relative rates of racemisation in enamel have been shown to display broadly similar trends (Dickinson et al., 2019) to those reported for free amino acids in an aqueous solution (Asp>Phe>Ala>Glu>Val; Smith and Evans, 1980). This broad trend was also observed in all the taxonomic groups studied here; for example the racemisation was lower in Glx (a slower racemising amino acid) than Asx for nearly all samples (Figure 4). Where near complete racemisation was observed in Glx (D/L ~ 1), Asx concentrations were too low to obtain reliable racemisation data (e.g. rhinocerotid samples TR5, TR7, TR8- Figure 4A and equid sample TE15- Figure 4B), consistent with heavily degraded protein trapped within the intra-crystalline fraction (Penkman et al., 2013). Two samples (rhinocerotid TR12 and bovid MB1; already discussed in section 3.1.1 for deviations observed in their racemisation data) had relative rates of racemisation that deviated from this trend, with either similar or higher extents of racemisation observed in Glx to Asx, potentially indicative of contamination (Willerslev et al., 2007) and/or mineral diagenesis (Preece and Penkman, 2005).
3.1.3. Peptide bond hydrolysis
Peptide chain hydrolysis can be investigated from IcPD data by calculating the relative percentage of free amino acids (%FAA) and allows an additional corroboration of extent of protein degradation. Hydrolysis is however considered a less predictable protein degradation mechanism with respect to rates (Walton, 1998), partly because its calculation compounds the errors derived from the low masses and volumes involved (rather than the cancellation achieved in D/L values) which occur during the calculation of concentrations (Powell et al., 2013). In addition, FAA formation can be affected by other degradation mechanisms such as decomposition (e.g. Ser decomposition to Ala; Bada et al., 1978) and lactam formation in Glu (Walton, 1998). %FAA therefore results in less precise data than racemisation (e.g. Penkman et al., 2013), which can be observed in the higher subsample variability (Figure 3). Nevertheless, peptide bond hydrolysis remains a useful marker for general protein degradation, and where closed-system behaviour is maintained, different protein degradation pathways, such as hydrolysis and racemisation, should be closely related (Penkman et al., 2013).
Most amino acids (with the exception of Ser and Asx, Stephenson and Clarke, 1989; Takahashi et al., 2010; Demarchi et al., 2013b) are not able to racemise when bound within a peptide chain (Smith and Evans, 1980; Mitterer and Kriausakul, 1984), requiring more conformational freedom, either as the terminal amino acid or completely free. Where closed-system behaviour has been maintained and degradation markers only result from endogenous protein, increasing degrees of racemisation should be correlated to increasing degrees of hydrolysis, broadly supported by this data (Figure 5; SI Figs. 5–8). In general, for Ala, Val and Phe (SI Figs. 5–8), an increase in free amino acid correlated with increased racemisation in all taxa, whilst for Asx (SI Figs. 5–8) and Glx (Figure 5), in the suid, equid and bovid, highly racemised samples resulted in a lower percentage of free amino acids (e.g. suid samples TS4, TS5 and equid samples TE8, TE15, Figure 5). The lower concentration of free amino acid in these samples may be as a result of decomposition typical of later stages of degradation. Differences in degradation patterns were observed for the rhinocerotid data in comparison to suid, equid and bovid, discussed further in section 3.2.
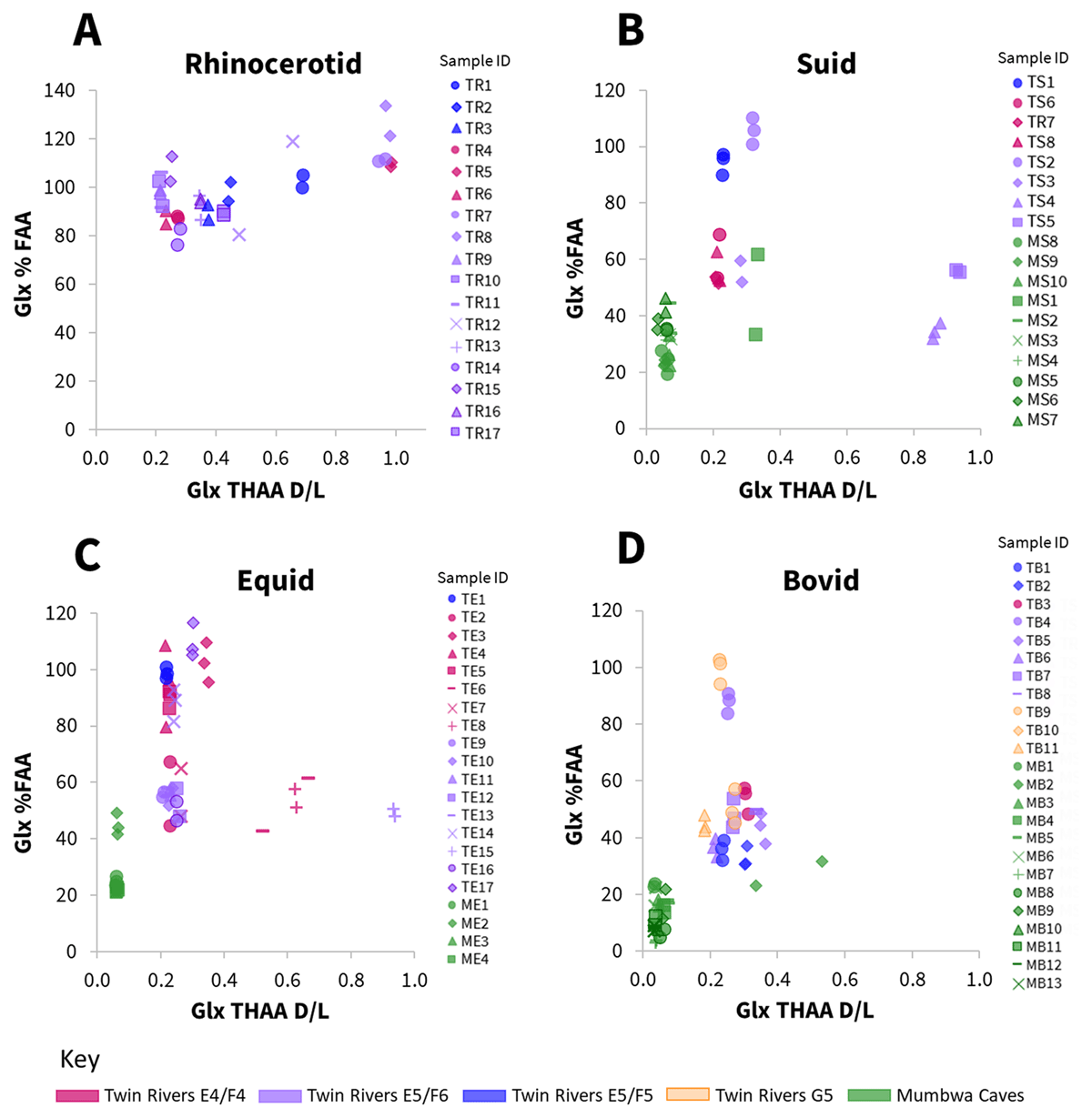
Figure 5
Relationship between racemisation values (THAA D/L) in Glx vs the percentage of free Glx, for subsample replicates, in four taxonomic groups of fossil enamel samples (A – rhinocerotid; B – suid; C – equid; D – bovid). Mumbwa Caves samples were all excavated from one deposit (Barham, 2000). Twin Rivers samples were excavated from four areas within the A Block cave passage (Barham, 2000).
3.1.4. Serine degradation
During protein diagenesis, dehydration of serine results in the production of alanine (Bada et al., 1978); in a closed system, [Ser]/[Ala] (the concentration of serine divided by the concentration of alanine) can therefore be used as a measure of protein decomposition. As discussed in section 3.1.3, where closed-system behaviour is maintained, a relationship would be expected between different protein degradation pathways. As for hydrolysis, the decomposition of serine was closely correlated with racemisation. [Ser]/[Ala] rapidly decreased when Glx THAA D/Ls were low (~0.0–0.2) and slowed after Glx D/L ~ 0.2 (Figure 6), indicating the retention of degradation products within a closed system (Penkman et al., 2008). For four samples, large differences were observed between subsample replicates (TR12- Figure 6A, TE6 and TE10- Figure 6C and MB2- Figure 6D), with one falling within and one outside of the trend); these divergent subsample replicates were the same ones which showed divergent racemisation data (Figure 4), indicating consistent subsample variability across multiple degradation pathways.

Figure 6
Covariance plots of average Glx racemisation vs serine decomposition in four taxonomic groups (A – rhinocerotid; B – suid; C – equid; D – bovid). No rhinocerotid samples were excavated from Mumbwa Caves, which all came from one deposit (Barham, 2000). Twin Rivers samples were excavated from four areas within the A Block cave passage (Barham, 2000).
3.1.5. Concentration and Composition
Concentration and relative composition are useful additional tools for closed-system behaviour assessment. As both vary with the extent of degradation, they are therefore best interpreted in combination with additional protein degradation markers. For example, low amino acid concentrations could result from highly degraded protein within a closed system but may also arise from leaching of material out of an open system. Similarly, relative amino acid composition can help to indicate when contamination (e.g. from dentine or exogenous sources) may be present in a sample (Dickinson et al., 2019), but profiles will change as a result of degradation (e.g. Ser dehydration to Ala; Bada et al., 1978). Exemplified by the rhinocerotid and equid data (Figure 7; all data shown in SI Figs. 9–12), the low THAA concentration and very high relative proportion of Gly observed for rhinocerotid sample TR8 (Figure 7A) indicated heavily degraded protein, corroborated by the high levels of racemisation (Figure 4), hydrolysis (Figure 5) and serine decomposition (Figure 6). However, for equid sample TE6 (Figure 7B), the low concentration and atypical composition (high relative proportions of Leu and Ile) were observed in combination with highly variable (off-trend) racemisation (Figure 4), hydrolysis (Figure 5) and serine decomposition (Figure 6) data, indicating open-system behaviour and/or contamination by exogenous protein.

Figure 7
Top – average total amino acid concentration, bottom – relative amino acid composition, for each enamel sample (A – rhinocerotid; B – equid). Boxes highlight low concentrations and atypical composition profiles.
3.1.6. Summary of identified outliers
Eight enamel samples showed evidence of possible open-system behaviour and/or contamination. This was observed from a combination of large, consistent, replicate variation across multiple degradation pathways, relative rates of racemisation and protein concentration and composition. While all the data is reported in the SI, these eight samples (TR12 (rhinocerotid); TS6 (suid); TE2, TE6, TE10, TE12 (equid); MB1, MB2 (bovid)) were removed from the final dataset used for each taxonomic group in the following sections. The remaining 72 samples, which appeared to adhere to closed-system behaviour (sections 3.1.1–3.1.5), were used for subsequent assessment of taxonomic effects (section 3.2), the fossils’ suitability for an enamel South-Central African amino acid geochronology and investigation of site formation processes (section 3.3).
3.2. Taxonomic effect
A taxonomic effect of amino acid racemisation rates has previously been described in a range of taxa (e.g. King and Hare, 1972; Bright and Kaufman, 2011; Ortiz et al., 2013), including within enamel (Dickinson et al., 2024). Rates of degradation, including racemisation, are influenced by a protein’s primary amino acid sequence (Mitterer and Kriausakul, 1984); as this differs between taxa in the enamel proteome, a taxonomic effect in racemisation may be expected. To investigate whether family level differences could be observed in this dataset, IcPD degradation trends were investigated. For racemisation (Figure 8A and SI Fig. 14) and serine decomposition (Figure 8B), no family level taxonomic differences were observed in their overall degradation trends, although the lack of chronological control in the dataset (see section 3.3 for discussion) meant that it was not possible to determine if their relative rates between taxonomic groups were different. There was however an observable difference in the degradation trend of rhinocerotid hydrolysis in comparison to suid, equid and bovid (Figure 8C&D; SI Figs. 15–18). For the rhinocerotid samples, very high levels of hydrolysis were observed over a broad range of racemisation values in most amino acids (e.g. >80% FAA between 0.2–1.0 D/L, Figure 8D, SI Figs. 15–18). However, for the other three taxonomic groups (suid, equid, bovid), a different peptide chain hydrolysis (%FAA) degradation trend was observed, with a potential decrease in hydrolysis at higher racemisation values (Figure 8D, SI Figs. 15–18). These apparently lower extents of hydrolysis in the more degraded samples likely indicate the loss of free amino acids through additional degradation mechanisms. Interestingly, in many CaCO3 biominerals, Glu lactam formation in the free amino acid fraction (Vallentyne, 1964; Walton, 1998) has been inferred from low Glu FAA concentrations, with %FAA plateauing at ~50% (e.g. Penkman et al., 2008; Crisp et al., 2013a; Tomiak et al., 2013). This trend however is not observable in this dataset. This may suggest a fundamental difference in protein degradation mechanisms between these types of biominerals or could indicate the additional biphasic separation step for enamel (as a CaPO4 biomineral; not included in CaCO3 methodology), could be influencing the observable degradation products (Baldreki, 2024).
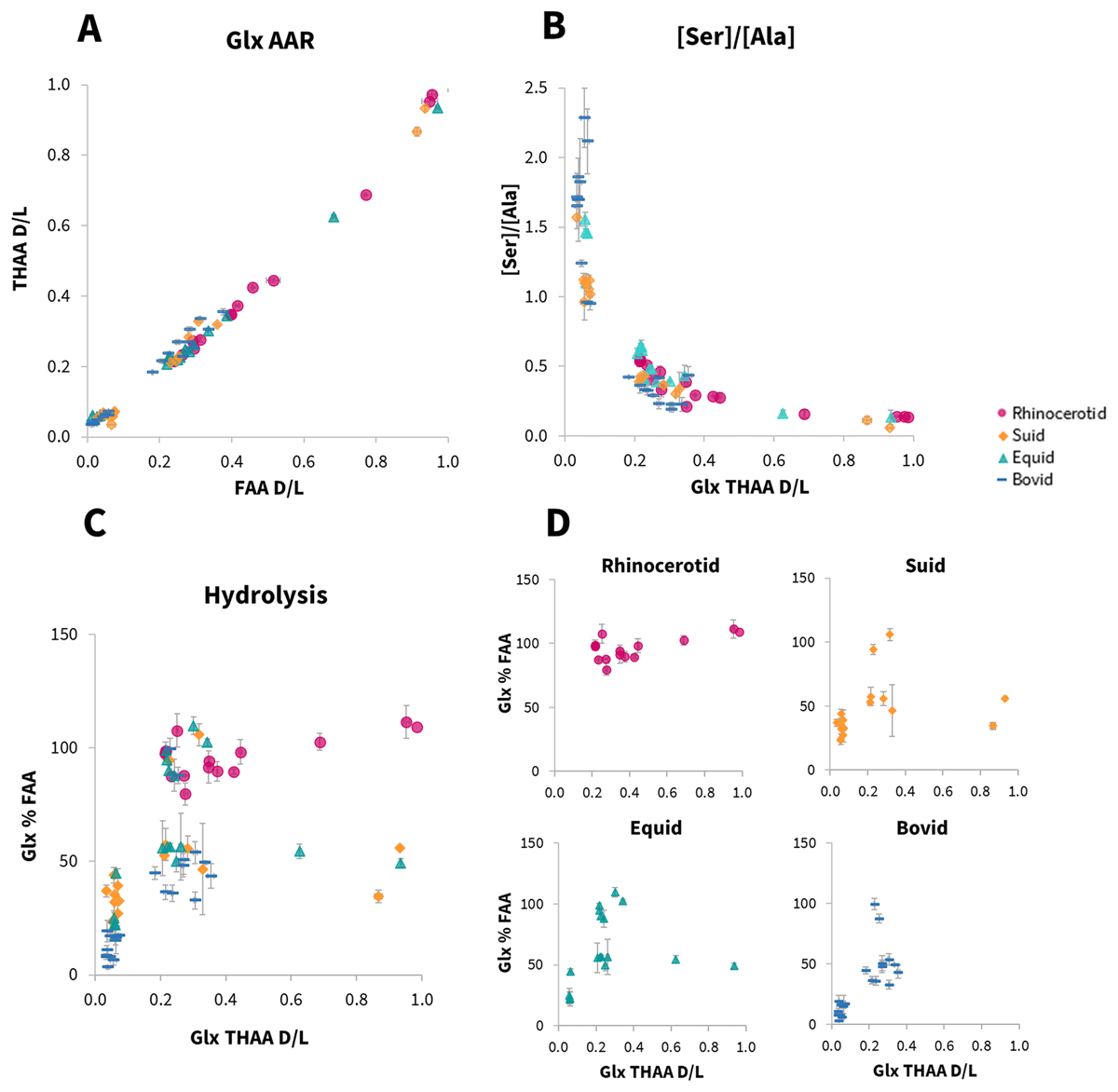
Figure 8
Investigation of family level taxonomic effect on the intra-crystalline protein degradation trends in rhinocerotid, suid, equid and bovid enamel samples. A – relationship between the average free (FAA) vs total hydrolysable (THAA) Glx racemisation values (D/L). B – relationship between serine decomposition vs total hydrolysable (THAA) Glx racemisation values (D/L). C – overlay of the relationship between percentage free (%FAA) Glx vs total hydrolysable (THAA) Glx racemisation values (D/L). D – individual taxonomic plots from C.
The apparent differences in family level taxonomic degradation trends observed in some of the IcPD parameters means that we therefore recommend building enamel amino acid geochronologies within family level taxonomic groups (at a minimum).
3.3. Assessment of site chronologies and depositional histories
IcPD analysis of fossil biominerals can provide useful direct dating information, enabling regional comparisons between sites, as well as within-site chronologies. It is not uncommon for sites, especially caves, to have experienced complicated depositional histories. Direct dating using a relative technique (such as IcPD) can be used to investigate individual site depositional histories (e.g. whether events such as bioturbation and erosion may have occurred), to allow a more robust understanding of each site’s sedimentary processes.
3.3.1. Mumbwa Caves
Relatively low levels of racemisation were observed for all samples from Mumbwa Caves (Figure 8), consistent with the site’s relatively young age (a maximum of >172 ka). One notable exception (suid MS1, Figure 9 A) had considerably higher D/L values for the majority of amino acids analysed (e.g. Asx Figure 9A). As the stratigraphic information for all Mumbwa Caves samples had been lost, it is not possible to relate the extent of racemisation with relative age, and therefore it is difficult to assess the likelihood of this being a genuine marker of depositional time. It is worth noting that this site has evidence of hearths in multiple archaeological horizons. Any tooth in close proximity to fire during its depositional history, could result in accelerated protein degradation and the extent of racemisation no longer reflecting primarily a signal of time (e.g. Brooks et al., 1990; Crisp, 2013b). Previous work to identify signatures characteristic of heating has been undertaken on intra-crystalline protein in ostrich eggshell (where high %Glx composition vs Asx D/L and/or low %Ala composition vs Ala D/L observed were indicative of heating; Crisp, 2013b); however we have too little comparative data in this study to investigate any characteristic heat-induced degradation trends in enamel, and it is therefore not currently possible to assess this in any of the samples analysed here, including suid sample MS1. With a range of racemisation values observed, this data does however provide an intra-site relative chronology for the material analysed.
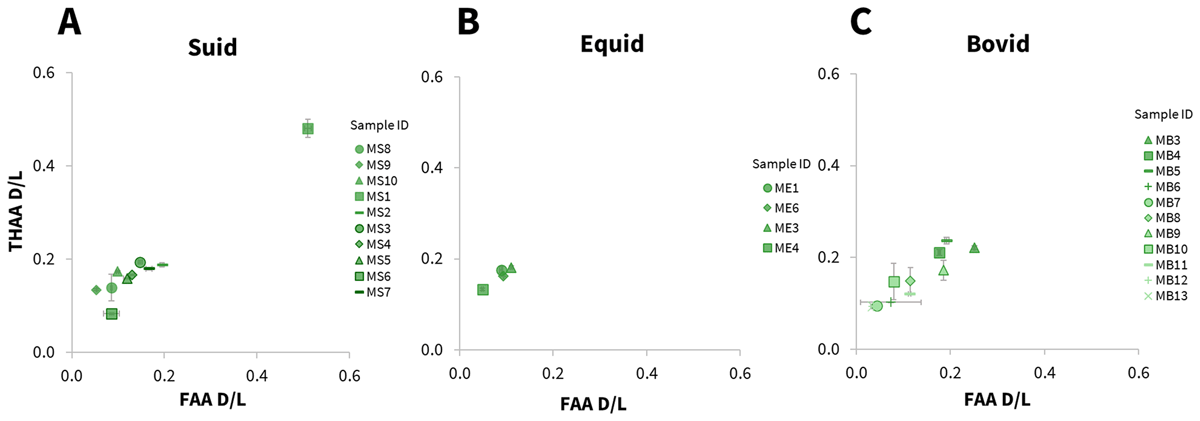
Figure 9
Relationship between the average free (FAA) vs total hydrolysable (THAA) Asx racemisation values (D/L) in three taxonomic groups of enamel samples present at Mumbwa Caves (A – suid; B – equid; C – bovid).
3.3.2. Twin Rivers
As stratigraphic information was available for the samples from Twin Rivers, the relationship between the extent of protein degradation and excavation depth could be investigated. Only in one excavation cavity (E5/E6), for two of the four taxonomic groups were there enough samples to investigate the relationship between the extent of racemisation and excavation level (Figure 10; SI Fig. 19). Whilst each excavation level is likely to be time-averaged, for both rhinocerotid (Figure 10A) and equid (Figure 10B), the sample with the lowest extent of racemisation observed for each excavation level (and therefore assumed not to be reworked) increased with depth. This is concordant with the working hypothesis of sediment deposition having washed into the sloping cave passage. There were however a number of levels displaying a large spread of racemisation values (e.g. rhinocerotids in E5/E6 level 1 (Figure 10A) and equids in E5/E6 level 4 (Figure 10B; SI Fig. 19), beyond what might have been expected from the horizontal excavation stratigraphy. Given these fossils showed closed-system behaviour (supported by the conclusions of section 3.1), and with no archaeological evidence of heating in A Block at Twin Rivers, we are left with the interpretation that the cave passage may have greater complexity within its depositional history, including considerable reworking of the fossils/sediments within the stratigraphy. The evidence of potential for bioturbation by tree root systems (Figure 2) provides one clear mechanism for this reworking, in addition to the known complexities of cave site taphonomies (e.g. Brain, 1983; Adams et al., 2007; Thompson, 2010; Val et al., 2015).

Figure 10
Average total hydrolysable amino acid (THAA) racemisation in Glx plotted against excavation level (1 = top 5 = bottom) in one excavation cavity (E5/E6) for rhinocerotid (A) and equid (B) samples. Error bars represent the standard deviation about the mean for subsample experimental replicates.
Whilst the lack of independent evidence of age precludes the ability to use this site to build an amino acid geochronology for the South-Central African region, the IcPD analysis has provided key information to allow a more informed interpretation of the site, by providing relative dates on the fauna for direct comparison. It is also clear that the faunal material covers a range of geological time at this site, with a wide spread of racemisation values observable in the majority of species (Figure 10, SI Fig. 19).
3.3.3. Between-site comparison
Whilst investigation of Mumbwa Cave’s stratigraphic adherence to chronological deposition was not possible, comparison to Twin Rivers, known to be an older archaeological site, was possible. With the exception of one relatively degraded suid sample (MS1; Figure 11), all samples from Mumbwa Caves had racemisation values lower than from Twin Rivers, in agreement with their younger age from previous dating. Interestingly, the published dates from both sites suggest there could be a temporal overlap between the sites (at ~170 ka); only in the suid materials is a possible overlap evident (Figure 11).
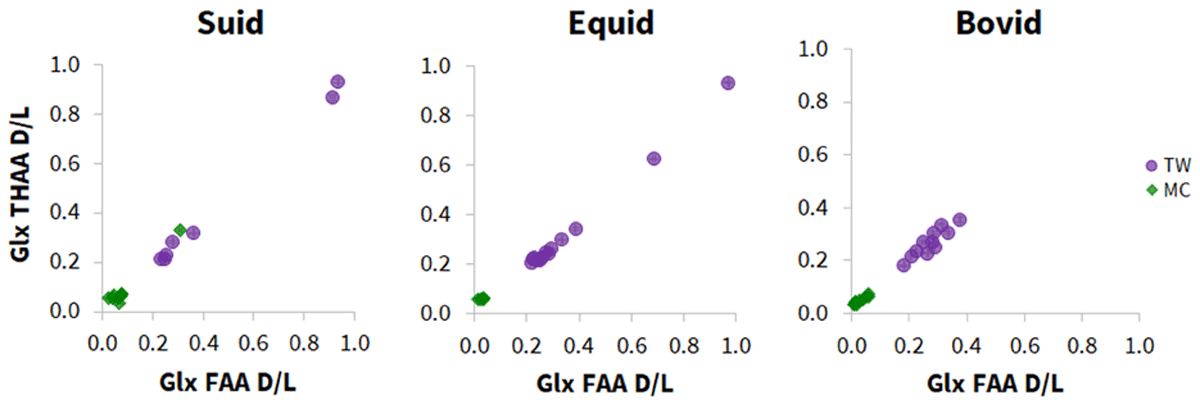
Figure 11
Relationship of the average free (FAA) vs total hydrolysable (THAA) Glx racemisation values (D/L) in the three taxonomic groups (suid, equid, bovid) of enamel samples present from both Mumbwa Caves (MC) and Twin Rivers (TW).
As the contextual information is not known for the fossils from Mumbwa Caves, one explanation is that the majority of fossils analysed from Mumbwa Caves represent a younger selection from later in the cave sequence. This could also mean the one exception where there is overlap with Twin Rivers samples (MS1, Figure 11), is an accurate representation of time at ~170 ka, and not as a result of proximity to a heat source (as discussed in section 3.3.1). However, as the flowstone capping the topmost part of the Twin Rivers A block sequence provides a minimum age of ~170 ka, the fossils excavated below may be older. Further study is required to better understand this dataset, but despite this, the IcPD data enables relative age assignments for this set of samples.
Conclusion
Eighty fossil tooth enamel samples from two archaeological cave sites in the South-Central African region (Twin Rivers and Mumbwa Caves) were analysed for IcPD to investigate their use for amino acid geochronology. Seventy-two of these (90%) showed evidence of closed-system behaviour in their intra-crystalline protein fraction. The relative extent of racemisation between sites was consistent with previous dating information. At Twin Rivers, a potential trend between the extent of racemisation in the least degraded samples and excavation depth was observed, concordant with the working hypothesis of a sequential deposition within A Block. Post-depositional processes (including bioturbation by roots and the mixing of deposits on excavation) may account in part for the wide spreads of racemisation values within some individual excavation levels, although greater depositional history complexity is likely. Direct dating of the fauna allowed recognition of this complexity, critical for interpretation.
The unrecoverable contextual information of the Mumbwa Caves material, combined with the data complexities at Twin Rivers, limited investigation of taxonomic variability within the IcPD dataset of enamel. Whilst a more thorough investigation of the taxonomic effect was not possible from these sites due to the lack of stratigraphic control, one notable degradation pattern difference in peptide chain hydrolysis was observed in the rhinocerotid data, in comparison to the suid, equid and bovid data, leading to the recommendation that taxon-specific enamel AAGs are developed. Although it was not possible to calibrate a regional aminostratigraphy based on these sites, the IcPD data provided key information for site interpretation allowing relative age determination for both individual faunal samples and between-site comparisons. At Twin Rivers the broad range of racemisation values observed in Glx (0.2–1.0 D/L), one of the slowest racemising amino acids, indicated the likely presence of fauna over a large portion of the Pleistocene, highlighting enamel’s potential for building Quaternary AAGs in warm regions, such as South-Central Africa. This study also provides a case study for the cautious approach needed when interpreting archaeological cave sites, and the value of direct dating material.
Data Accessibility Statement
Data in this study has been included in the supplementary information and will be available on the NOAA data repository upon publication: https://www.ncei.noaa.gov/pub/data/paleo/aar/
All teeth/fragments sampled in this study will be archived in Livingstone Museum, Zambia.
For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising for this submission.
Additional Files
The additional files for this article can be found as follows:
Acknowledgements
We would like to thank Livingstone Museum and the Zambian National Heritage Conservation Commission for their help and support. We would also like to thank Sheila Taylor and Samantha Presslee for technical support, and Lucy Wheeler for their valuable comments on the manuscript.
Funding Information
This work was supported by the Natural Environment Research Council [NE/S00713X/1 and NE/S010211/1]. Research at Mumbwa Caves was funded by the LSB Leakey Foundation, the British Academy, the National Geographic Society and the Prehistoric Society and the Natural Environment Research Council. The excavations at Twin Rivers in 1999 were funded by the LSB Leakey Foundation.
Competing Interests
The authors have no competing interests to declare.
