Poultry meat can be contaminated during many stages of the slaughtering process. It is known that there are critical points in the slaughter process and sites in the slaughterhouse environment which are the main sources for contamination of chicken carcasses (28) and remain so despite some environmental hygienic measures being used. In this context, keeping microbial growth under control is of critical importance and improves the shelf life and quality of the carcass and derived products, especially in commercial poultry processing (14).
There have been many studies on monitoring, detection and counting to identify the source of bacterial contamination, as these research topics have long been recognised as essential for commercial poultry production (6, 7, 22). The level of contamination of poultry carcasses is directly related to many factors, including how live birds are kept at the pre-slaughter stage and cross-contamination between the processing equipment and the processing environment. The studies conducted so far have mostly been carried out for the detection and/or quantification of certain pathogens.
It is crucial to fully reveal the changes in the microbiome during processing. This will provide insight into how to reduce the rate of microbiological contamination of the final product by maintaining and developing an effective sanitation programme (10, 24). Among all contamination sources, processing chilling water, scalding water, contact surfaces and the airborne microbial load have been considered as the primary ones cross-contaminating carcasses and affecting the microbial distribution in the final product (21).
Limited data is available for determination of microbiota diversity through the different stages of the poultry slaughtering process and the surrounding environment. Although many studies which mainly deal with detection of different microbial communities have shown that there is a quantitatively significant decrease in microbial load after the storage stage in poultry slaughtering operations, the current metagenomic studies have provided an important result in terms of demonstrating broad microbial diversity. Another important aspect that should be considered is that these bacteria may revive from the dormant form to the form in which they can grow, especially under insufficient cooling or unsuitable storage conditions. Therefore, not only the airborne microorganisms, but also those that were undetectable in the previous stages could reach an environment with suitable conditions and increase in number. In this context next-generation sequencing (NGS) platforms perform sequencing of millions of small DNA segments in parallel. The use of NGS provides the opportunity to determine relatively low-abundance taxa and investigate their redistribution on chicken carcasses during the slaughter process (4). The aim of this study was to determine the diversity of the bacterial community in chicken carcasses and environmental sources at various stages along the poultry slaughter line by using NGS.
Chicken carcass samples and slaughter-line environmental samples were collected between September and November in 2021. The poultry slaughterhouse was a vertically integrated operation where approximately 4,500 birds are slaughtered per hour. After the birds are stunned by electric shock (100V, 3–5 s), they are slaughtered manually using a knife. After bleeding for approximately 6–7 min, the birds are transferred to scalding tubs which contain water at 62–64°C for 180 s. The carcasses are defeathered automatically, eviscerated manually by the personnel and then washed before the chilling stage. The chilling process in the slaughterhouse is carried out using immersion chilling in water in two steps.
Rinsate samples from whole chicken carcasses from four different stages of the slaughter process were analysed. These stages were after defeathering (AD), after evisceration (AE), after water chilling (AC) and after cooling to storage temperatures (AS, immediately before shipping) (Fig 1.). On each monthly visit, three chicken carcasses were randomly collected at each stated slaughter stage, making 36 chicken samples in total. Whole chicken carcasses were sampled using a chicken rinsing method: a carcass from a particular slaughter process stage was placed in a sterile stomacher bag and rinsed by hand with 500 mL of buffered peptone water (BPW – Oxoid, Basingstoke, UK) for 2 min immediately after collection. The rinsate sample was then transferred into a sterile borosilicate glass bottle. Contact surfaces of the processing areas (the table, floor and cutting utensils) were randomly swabbed and the swab samples pooled to represent the environmental surface (ES) samples. Sterile cotton swab kits were prepared using BPW and 10 × 10 cm2 surface areas were sampled in the processing hours. Also, samples of scalding tank water (SW; n = 3) and chilling tank water (CW; n = 3) from 10 cm below the surfaces of the tanks were collected into sterilised 600 mL jars. Personnel hand (PH) samples (five personnel hands on each visit, n = 15) were also collected using the swab kits mentioned above. For hand sampling, cotton swabs were rubbed onto a 5 × 5 cm2 surface of the right hand of each person including the palm, fingers and fingernails.
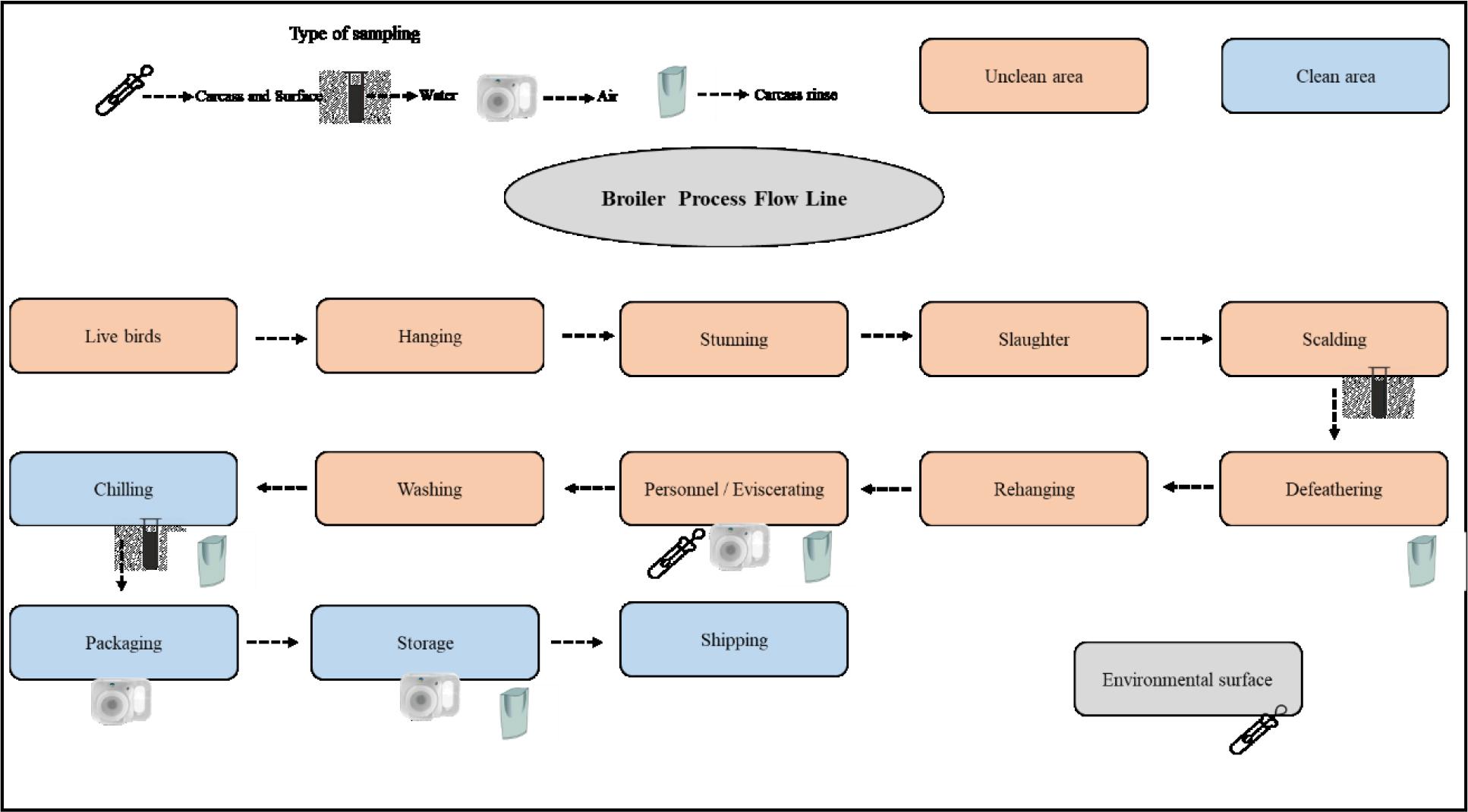
Sampling points in process line
Air samples of the evisceration (EA), packaging (PA) and storage (SA) rooms were obtained using an active air sampling device (Air Ideal; BioMérieux, Marcy l’Étoile, France) which measures 190 L of air with an impact speed <20 m/s and then blows that air onto a medium. For this purpose, Plate Count Agar (PCA; LabM, Potters Bar, UK) was used to determine the diversity of culturable aerobic mesophilic bacteria. The air sampler was positioned on an appropriate surface, which was the centre of a wall of each room.
All the samples were transferred to the laboratory in a cold chain and analysed within 2 h. The plates from air sampling were incubated at 30°C for 48 h, and colonies were washed with sterilised 0.09% NaCl from the surface of the agar plate. The resulting homogenates of air samples and swab samples (500 μL for each) were placed in sterile Eppendorf tubes and then processed for DNA extraction.
Bacterial DNA extraction with cetyltrimethylammonium bromide was performed according to the classical method described by Barouni et al. (2).
To determine the microbiota of poultry carcasses and environmental samples along the slaughter line, the V3–V4 regions of the 16S rRNA gene were amplified with the MiSeq341F (5’-TCG TCGGCAGCGTCAGATGTGTATAAGAGACAGCC TACGGGNGGCWGCAG-3’) and MiSeq805R (5’-GTC TCGTGGGCTCGGAGATGTGTATAAGAGACAGG ACTACHVGGGTATCTAATCC-3’) universal primers (11), which covered an approximate 460-base-pair region. The 5’-ends of the primers are the Illumina sequence adapters and the 3’-ends amplify the V3–V4 region of the 16S rRNA gene.
A PCR was performed using a KAPA HiFi HotStart ReadyMix PCR Kit (Kapa BioSystems, Wilmington, MA, USA). The steps were initial denaturation at 95°C for 3 min; 25 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 30 s; and final extension at 72°C for 5 min.
Amplified products were purified using the Agencourt AMPureXP Kit (Beckman Coulter, Brea, CA, USA) according to the manufacturer’s instructions. A second PCR reaction was performed for adapter sequences allowing binding of PCR products to specific oligobinding sites. The selected product was a Nextera XT Kit (Illumina, San Diego, CA, USA). The thermal conditions were 95°C for 3 min; 8 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s; and 72°C for 5 min. Following the PCR, the amplified products were purified.
A KAPA Library Quantification Kit (Kapa Biosystems) was utilised for quantification of the libraries using the Eco Real-Time PCR System (Illumina). Following the cluster generation, the templates were sequenced using the Illumina NovaSeq 6000 System (Illumina). To generate high-quality sequence data, the DNA fragments from clusters were sequenced first from one end and then from the opposite end. After the sequencing, raw data was demultiplexed by means of indices.
Quantitative Analysis in Microbial Ecology (QIIME2) was utilised to plot Operational Taxonomic Units (OTUs) and to assess alpha and beta diversity of the sequenced clusters (3). The cut off value was a 95% identity level. Based on OTU quantities, Shannon and Simpson index values were computed to assess the alpha diversity of the samples. A (dis)similarity matrix was assembled using beta diversity calculated using the Bray–Curtis metric of the data, containing the similarity between each sample (rows) and every other sample (columns). To visualise the sample similarity based on that, the data were condensed to a lower dimensionality, or principal components, using the principal coordinate analysis (PCoA) ordination technique. The data having been condensed to the lower dimensions, the hierarchical clustering algorithm (HCA) was employed to find clustered data. The algorithm’s aim is to keep maximising the distance between the different clusters (inter-cluster distance) and minimising the distance between objects belonging to the same cluster (intracluster distance). The HCA was deployed to split data into groups with Ward’s method and Euclidean distances between principal coordinates. Use was made of R Statistical Programming Language version 4.0.4 for all statistical analyses (18).
Alpha diversity analysis was performed to represent the number of taxonomic groups, and the Shannon and Simpson diversity indices were calculated to determine the richness of species in the samples. A total of 1,234,899 qualified sequences were obtained in the samples. The higher Shannon index and lower Simpson index suggested a high diversity within the samples. The Shannon and Simpson indices demonstrated the AS and AC to be the stages which had the highest microbial abundance among the slaughtering stages. Among the environmental samples, the CW and PH samples were found to have the greatest microbial diversity (Table 1). The microbial alpha diversity index (Shannon) was observed to be increased at the AS stage, which indicates a more diverse bacterial community structure compared to the other stages. Table 1 displays the Shannon and Simpson values of the slaughtering stage and environmental samples.
Shannon and Simpson index values of the bacterial microbiome diversity along poultry slaughter lines by slaughtering stage and environmental sample type
| Samples | Slaughtering stages | Environmental | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | AE | AC | AS | ES | SW | CW | PH | EA | PA | SA | |
| Shannon | 6.040 | 5.562 | 6.347 | 6.847 | 5.331 | 4.427 | 6.474 | 6.422 | 3.732 | 2.452 | 2.852 |
| Simpson | 0.956 | 0.939 | 0.956 | 0.973 | 0.920 | 0.756 | 0.973 | 0.970 | 0.865 | 0.679 | 0.794 |
AD – after defeathering; AE – after evisceration; AC – after chilling; AS – after storage; ES – environmental surfaces; SW – scalding water; CW – chilling water; PH – personnel hands; EA – evisceration air; PA – packaging air; SA – storage air
Beta diversity was plotted as PCoA results displaying the relatedness of slaughtering stages and environmental samples (Fig. 2). Principal coordinate analysis transformed the data from the distance matrix into orthogonal axes, with the significance of each axis indicated by its eigenvalue, resulting in a three-dimensional representation of the data. The distance matrix was transformed into a set of orthogonal axes where the first axis (PC1; 51%) displayed the maximum amount of variation, the second the median (PC2; 17%) and third the minimum amount (PC3; 11%). Our beta diversity PCoA analysis depicted that microbiome diversity was lower in air samples (EA, SA and PA) and on environmental surfaces (ES). The eleven sample categories were grouped into four different segments with a similarity level of 62% and Bray–Curtis distance of 2.5. The first group included the AD, AE, AC and AS groups and formed cluster 1; the second group consisted of ES, SA, PA and EA, forming cluster 2; the third one contained CW and PH to create cluster 3; and the fourth one was only SW and simultaneously was cluster 4.
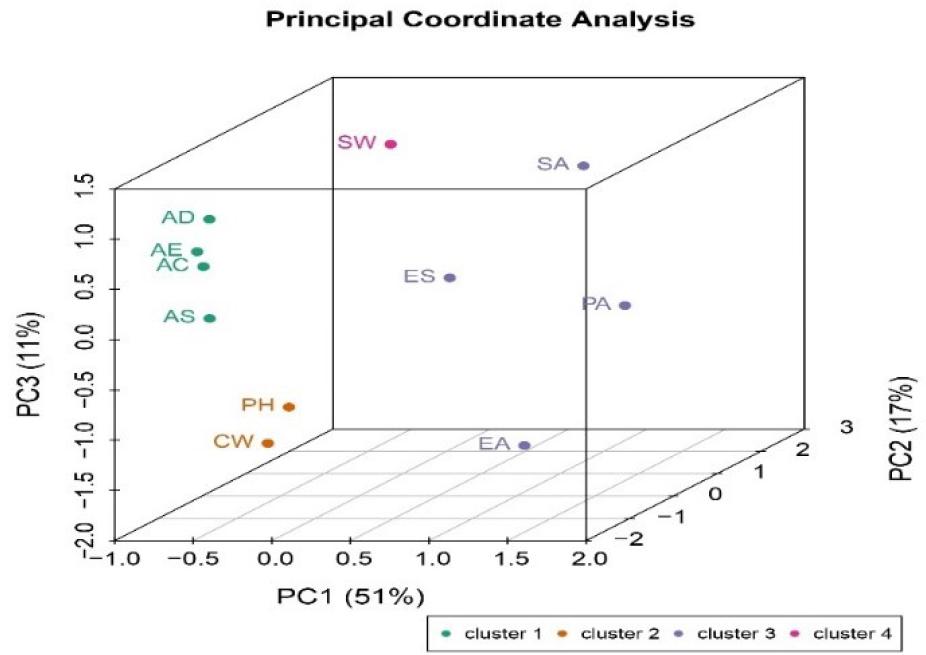
Principal coordinate analysis of beta diversity in the bacterial microbiome along poultry slaughter lines by slaughtering stage and environmental source
AD – after defeathering; AE – after evisceration; AC – after chilling; AS – after storage; ES – environmental surfaces; SW – scalding water; CW – chilling water; PH – personnel hands; EA – evisceration air; PA – packaging air; SA – storage air
The heatmap in the left part of Fig. 3 visualises the Spearman correlation coefficients between various pairs of variables, which measure the strength and direction of monotonic relationships without assuming normality or linearity. Notably, AS and AC (0.94) and AE and AD (0.84) exhibited strong positive correlations, and many others showed moderate positive correlations. In summary, among all sample groups, AS was determined as the stage with the most correlation with other sampling points, exhibiting such with AE, AD, AC and CW. Although the slaughtering stages were highly correlated with each other, the environmental sample CW, which had the most abundant genera according to the alpha diversity results, was weakly correlated with AS, AC, AE and AD.
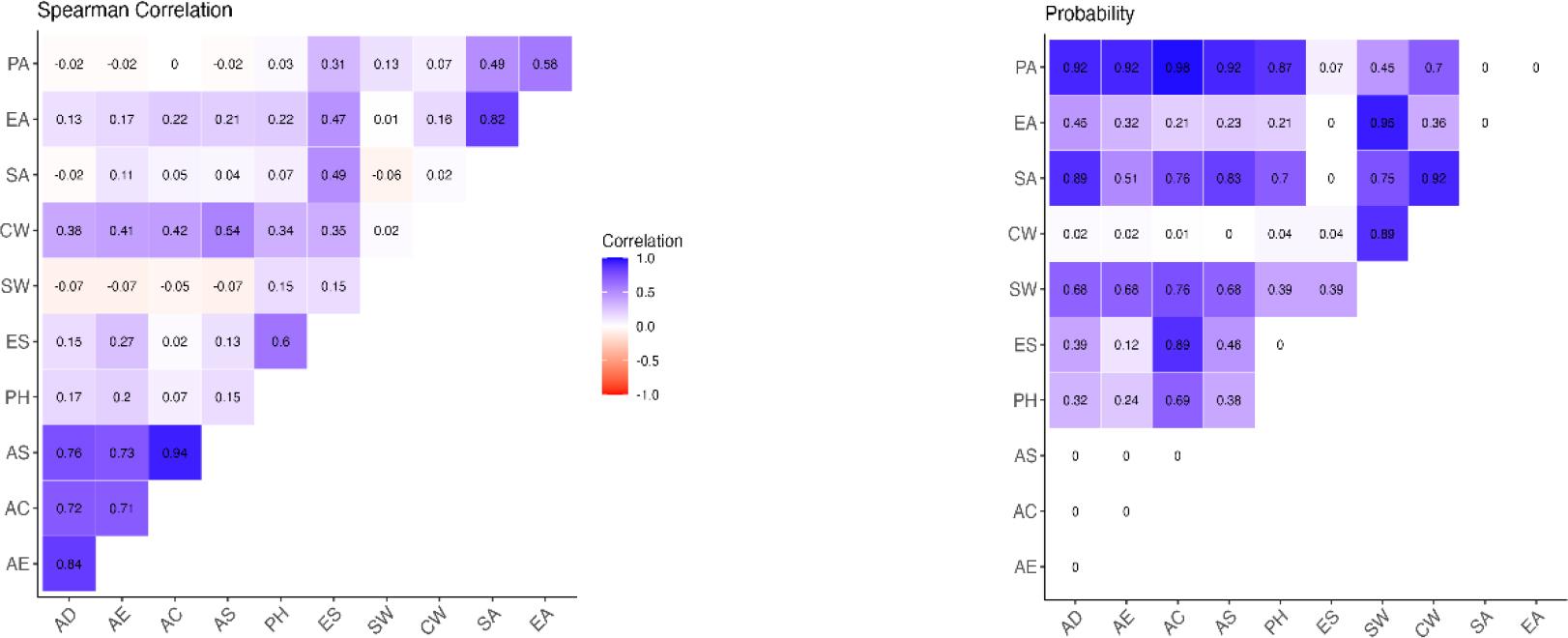
Correlation of beta diversity in the bacterial microbiome along poultry slaughter lines by slaughtering stage and environmental source. Darker blue and violet shades in the left figure represent stronger positive correlations, and red shades stronger negative correlations. Darker blue shades in the right figure represent correlations which are not statistically significant, and white shades statistically significant correlations
AD – after defeathering; AE – after evisceration; AC – after chilling; AS – after storage; ES – environmental surfaces; SW – scalding water; CW – chilling water; PH – personnel hands; EA – evisceration air; PA – packaging air; SA – storage air
The right part of Fig. 3 shows the results of hypothesis tests to determine whether each Spearman correlation coefficient was significantly different from zero. A P-value ≤ 0.05 indicated significance.
The microbial diversity of slaughtering stages and environmental samples is shown in Figs 4 and 5, respectively. The AS slaughter line point, which is the final station along the poultry slaughtering line, provided samples showing that the same microbial community that came from the AC stage remained dominant. Furthermore, the proportion of Acinetobacter (14.99%) increased, making it the dominant genus by a larger margin. Macrococcus and Enterococcus were also in high abundance, with 10.87% and 10.22%, respectively.
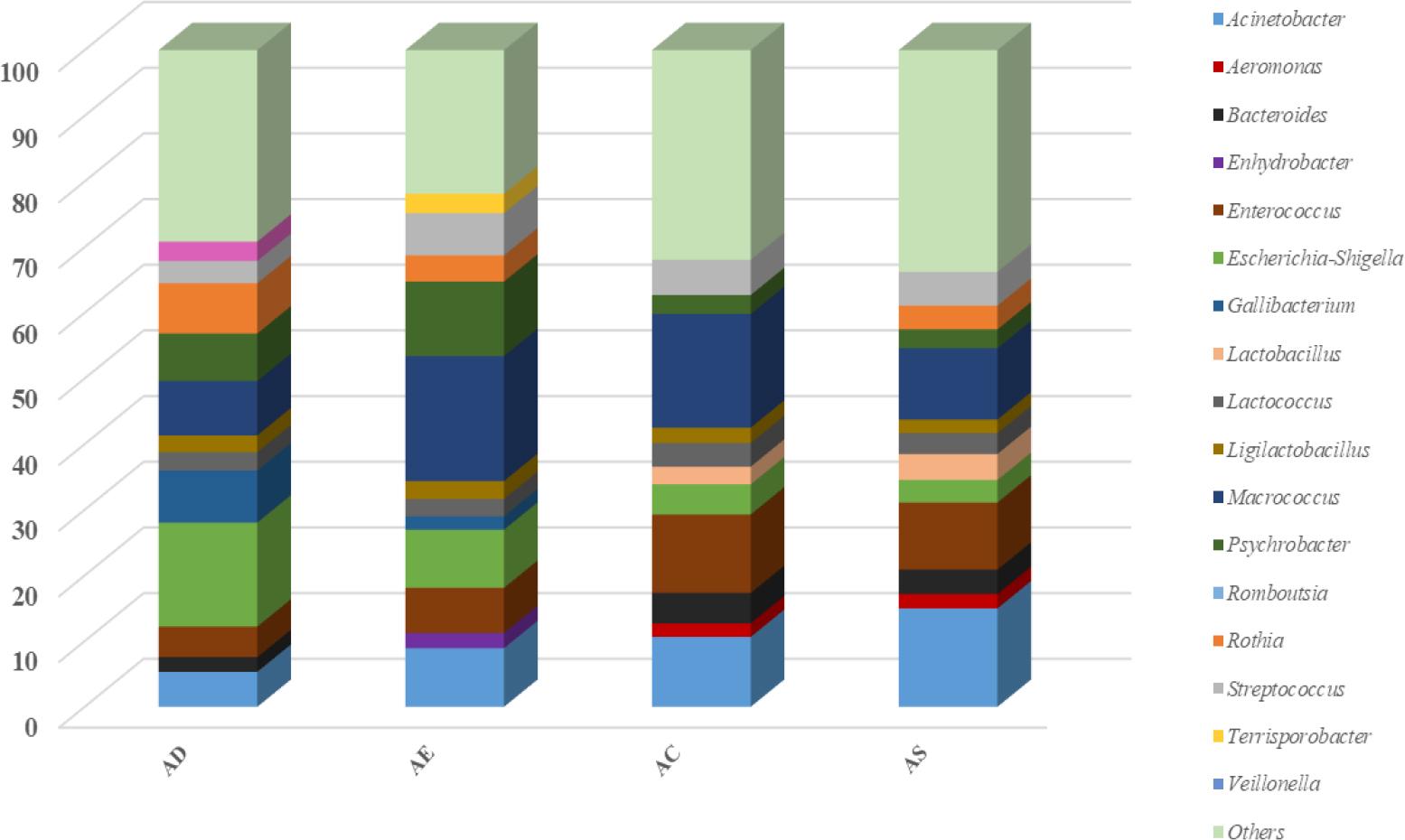
Microbial diversity along poultry slaughter lines by slaughtering stage
AD – after defeathering; AE – after evisceration; AC – after chilling; AS – after storage
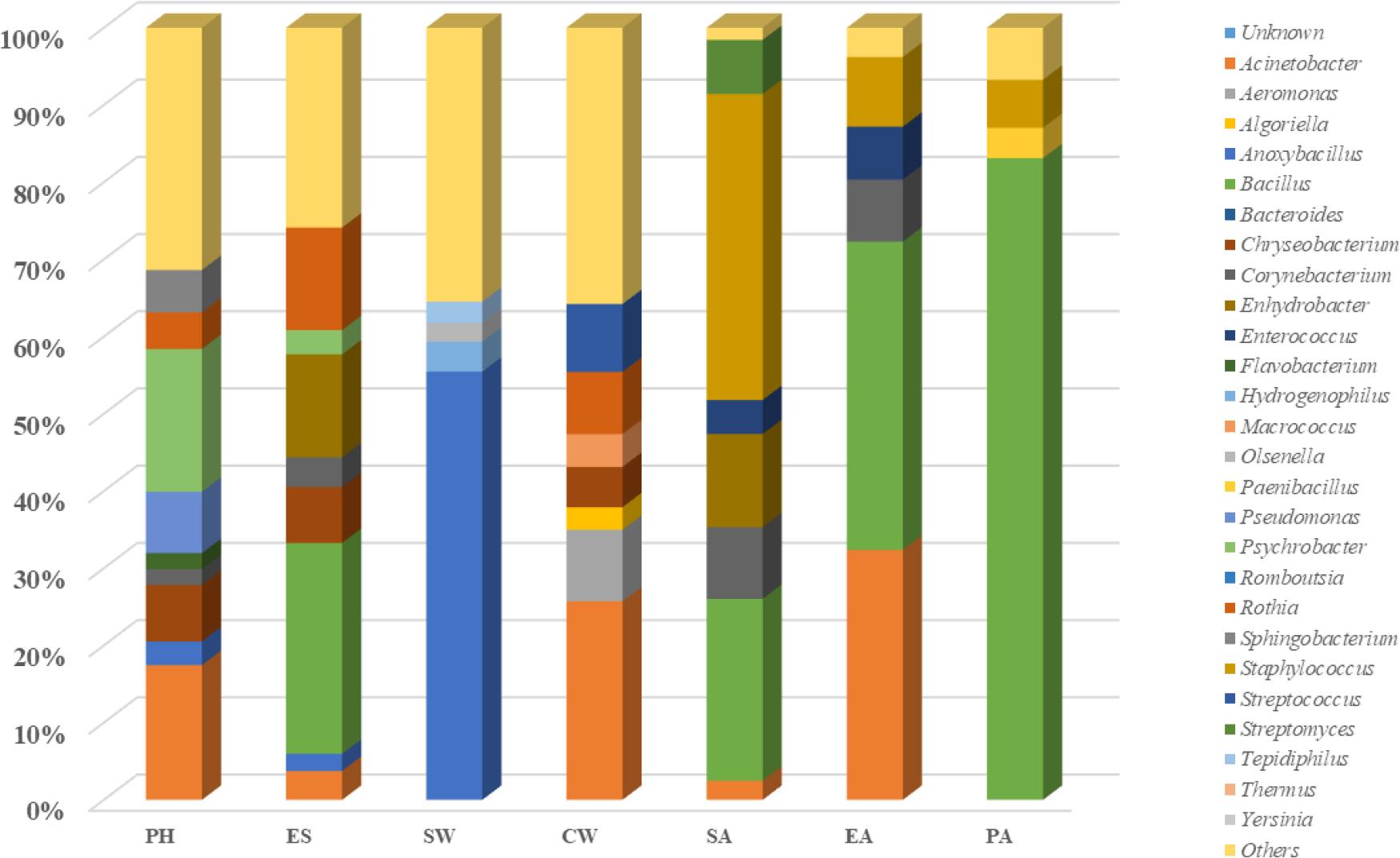
Microbial diversity along poultry slaughter lines by environmental source
PH – personnel hands; ES – environmental surfaces; SW – scalding water; CW – chilling water; SA – storage air EA – evisceration air; PA – packaging air
Of the environmental samples, CW had the highest microbial diversity, followed by PH. A comparable study by Song et al. (24) on yellow-feather chicken carcasses found a more complex microbial community after the chilling stage and in the environmental samples. The present study found microbiome diversity to be lower in air samples (EA, SA and PA) and on environmental surfaces (ES). The lower diversity in air samples was thought to be related to the sampling method, which limited what was detectable to only aerobic, mesophilic and culturable microorganisms. The relatively lower diversity in ES samples could be because of the use of decontaminating agents. Besides, the mean microbiome of all slaughtering stages (AS, AE, AD, AC) were mostly related to CW and PH according to beta diversity principal coordinate analyses. These results also indicated that the only washing stage, before chilling, was not effective, because the most microbially diverse and correlated stages were AC and AS. The chilling stage may also not suffice to decrease the microbial community because decontaminants are not used during the slaughtering stages. Similar inferences can be drawn from a recent study by Stella et al. (25), which compared different chilling strategies. The researchers suggested that mild decontaminating agents be used in the slaughtering stages and the chilling stage be considered a useful intervention point.
Our results suggest that the bacterial microbiome of chicken carcass samples and environmental samples was predominantly comprised of Proteobacteria, Firmicutes, Actinobacteriota and Bacteroidota. The same pattern has been previously reported by Kim et al. (10), Chen et al. (4) and Song et al. (24).
The relatively highly prevalent Escherichia-Shigella (15.83%) and Macrococcus (8.30%) species determined at the AD stage may have originated from the feather follicles. This result was in line with previous studies implying that these were major sources of carcass contamination (17, 30). This stage is known to have one of the highest rates of cross-contamination originating from contact surfaces, equipment, workers and other contaminated carcasses. Perez-Arnedo et al. (17) stated the defeathering stage as the major source of carcass contamination with Salmonella spp., E. coli, Campylobacter spp. and Staphylococcus aureus. The relative abundance of these genera and species at this stage could be attributed to the pluckers, which are particularly difficult to disinfect in a slaughtering process with very fast throughput; this machinery could be the major source of cross-contamination (8). These genera contain species which are human pathogens; therefore, minimising their presence should be considered critical for poultry processing.
The abundance of Macrococcus and some Enterobacteriaceae members like Escherichia-Shigella in the AE stage was considered a result of manual evisceration, which may lead to perforation of the caecal lining and the spread of caecal contents through the evisceration environment. The high prevalence in our result was consistent with the results of Roccato et al. (20), which implied that manually performed evisceration was a major factor for carcass contamination, and that it took place through workers’ hands or utensils. In a similar study, Perez-Arnedo et al. (17) stated that evisceration equipment may cause the perforation of the intestines and that intestinal content may be the source of the contamination of carcasses.
The high Acinetobacter content (10.65%) in the chilling tank was also observed to lead to an increase of this taxon on carcass surfaces in the AC stage. This genus was detected as one of the three most abundant at the chilling stage. This may be related to the high adhesion capability of microorganisms of this genus, because this characteristic has been stated to make it the primary contaminant for poultry processing environments (9). Savin et al. (23) similarly observed that the process waters were highly contaminated with Acinetobacter complex at different stages along poultry slaughter lines (36.5%). The other abundant genera after the chilling stage in the present investigation were Macrococcus (17.32%) and Enterococcus (11.95%), which retained and increased their dominance, respectively. A similar microbial genera pattern was reported by Zhang et al. (30), with Acinetobacter having the highest abundance, followed by Psychrobacter, Macrococcus and Comamonas, which were in greater than 2% abundance after chilling. Bacterial abundance and diversity after the chilling stage are generally related to the different chilling parameters, such as relative humidity and temperature profile, air velocity, and the mass and fat content of the carcass (17). The cooling technology and whether it relies on air or immersion systems are also known to be major factors and play critical roles in carcass contamination. Chen et al. (4) found that immersion chilling reduced the total viable count of E. coli and Campylobacter contamination on chicken carcasses compared with air chilling.
Environmental surfaces are the primary sources of cross contamination of pathogenic or spoilage bacteria in poultry slaughtering. During slaughtering, bacteria can be spread between carcasses by the equipment surfaces. The pooled ES sample swabbed from surfaces which are in contact with carcasses during defeathering and evisceration showed that Bacillus was the most abundant genus, followed by Enhydrobacter and Rothia. Bacillus spp. are ubiquitous microorganisms that may originate from many sources and their spores or cells are easily transferred in flocks and slaughterhouses. The adhesion of spores and resistance to external environments of some Bacillus species can lead to formation of biofilms which are highly resistant to disinfectants. In this way, cross-contamination via utensils may occur (1). Another abundant genus, Enhydrobacter, was reported as the most prevalent in biofilms on stainless-steel surfaces (16). Rothia, the other abundant genus in our results, is an opportunistic pathogen of animals and can cause disease in immunosuppressed humans. Alarmingly, in a recent study, Rothia nasimurium, which had been isolated for the first time from chickens, was found to be multiresistant (29).
Psychrobacter, Acinetobacter and Pseudomonas were the most abundant Gram-negative genera in the samples obtained from the PH samples. These genera are mostly known as food spoilage bacteria. Besides being common isolates from spoiled poultry meat products, Psychrobacter spp., with their psychrophilic nature, and Acinetobacter spp., which are resistant to many antibiotic groups, are generally the dominant species on poultry slaughter lines (9). The abundance of Pseudomonas spp. was noteworthy because the proteolytic and lipolytic activities of some of its species are the primary spoilage mechanisms in meat under cold storage conditions. The detection of these three genera in substantial amounts in the PH samples suggested that personnel-derived contamination can cause spoilage in the end product and also play a significant role in the spread of pathogenic microorganisms. Adequate preventive measures like separating the clean and unclean areas on the process line and training personnel in good hygienic practices can reduce the possibility of contamination during poultry slaughtering.
In our study, the chilling tank emerged as having the most diverse microbial community and may have been the primary source of carcass contamination. In the same way, Song et al. (24) found the most complex bacterial diversity in their chilling-tank samples. That the most diverse carcass contamination was observed in the storage stage can also be attributed to the failure of carcass cleaning to be sufficient in the chilling stage.
In SW, the very high abundance of Anoxybacillus (55.47%), members of which are spore forming, highly durable under high temperature and also very well known in the dairy industry as indicators of poor hygiene (19), was observed not to leave detectable carcass surface contamination at the defeathering stage.
It is known that there is a high level of microbial contamination in indoor air in poultry-processing facilities. Airborne microorganisms settle on an equipment surface microbial community and the personnel’s hands and cause contamination of the poultry carcass surface throughout all slaughtering stages (13, 27). The next-generation sequencing results from air sample isolates showed that Bacillus and Staphylococcus were the dominant species. A comparable study conducted by Lues et al. (13) to determine the microbial load in poultry slaughterhouse air samples found Bacillus cereus to be the most common bacterium. In another study conducted in air samples in different operational areas in a poultry slaughterhouse, Staphylococcus spp. was found to be in heavy presence in the evisceration and reception areas (5).
The most abundant species at the EA sampling point were Bacillus, Acinetobacter, Staphylococcus, Corynebacterium and Enterococcus. A pattern little different was observed in previous studies (13, 27). Among the slaughtering stages, evisceration is a critical point for contamination of air. Maharjan et al. (15) suggested that the microbial load of air samples decreased from high to low through the lairage, bleeding, evisceration, spin chilling, grading and packaging sections.
The PA and SA samples yielded abundant Bacillus spp. and Staphylococcus spp., respectively, and these species’ survival may be attributed to several factors of the ambient air. Temperature and relative and absolute humidity are the main factors that determine the survival of indoor airborne bacterial organisms such as Gram-positive bacteria, which are more resistant at intermediate relative humidity rates (approximately 50–70%) (26). The advantage Bacillus spp. have in airborne transmission over other airborne Gram-positive bacteria is their ability to sporulate under adverse environmental conditions. The higher presence of Staphylococcus spp. in the SA samples can be attributed to the more intense personnel activity in this area. It was also indicated by some researchers (5, 12) that the movement rate of people in an indoor place was one of the main factors for changes in air microflora. A limitation of our study in air samples was the methodology used, because plate count agar only detects culturable airborne microorganisms which have the ability to grow on PCA under mesophilic and aerobic conditions.
The metagenomic data obtained from poultry slaughtering stage and surrounding environment sampling showed that potential contamination and cross-contamination noted after storage in what was the final product mostly originated from prior slaughtering stages, and from chilling water and the hands of slaughter operation personnel as well. Previous studies based on quantitative analysis have always been consistent in their finding of a dramatic decrease at the chilling and storage stages, while the present study noteworthily found increased microbiome diversity at these stages, which demonstrates the effect of the physical conditions created on the microbial community pattern through redistribution. This diversification can result from cross-contamination from the previous stages. In addition, some microorganisms may survive and thrive under suitable environmental conditions which may be encountered in a poultry slaughterhouse. In this investigation, a microbiome-based insight was provided to implement a hurdle strategy for remedial actions by identification of the critical control points. In subsequent research, it would be helpful to combine metagenomic and quantitative data to better understand bacterial behaviours under different environmental conditions.