Figure 1

Figure 2
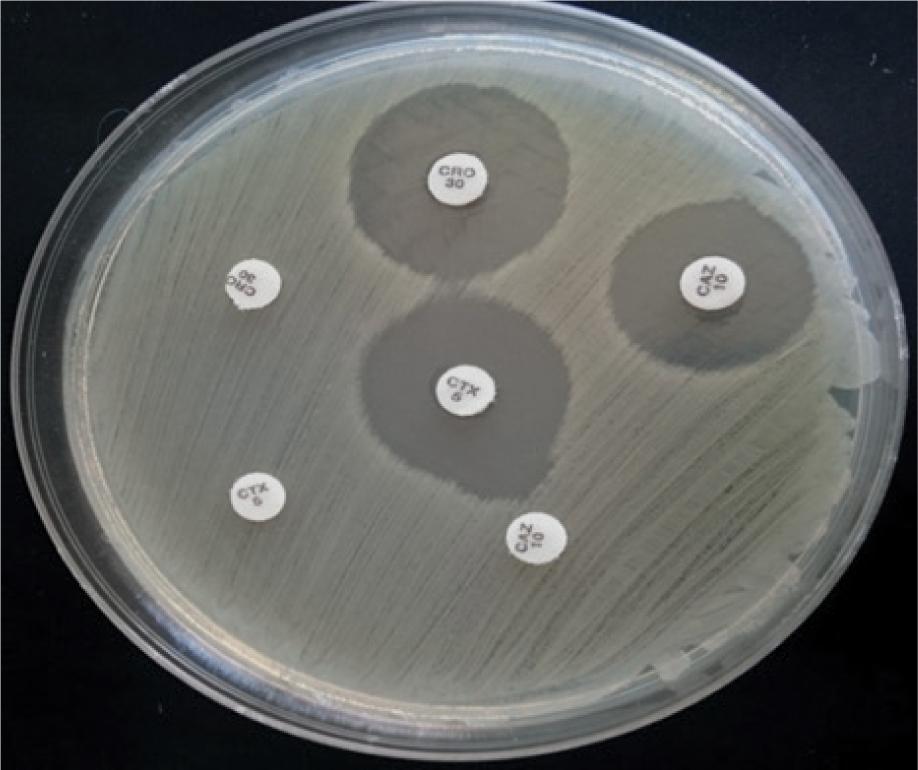
Figure 3
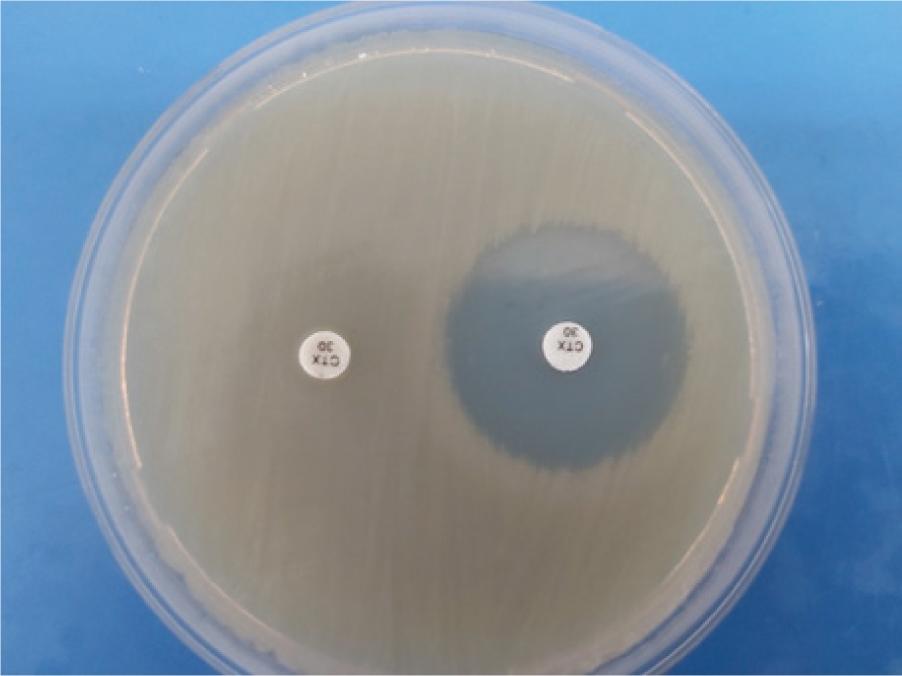
Figure 4
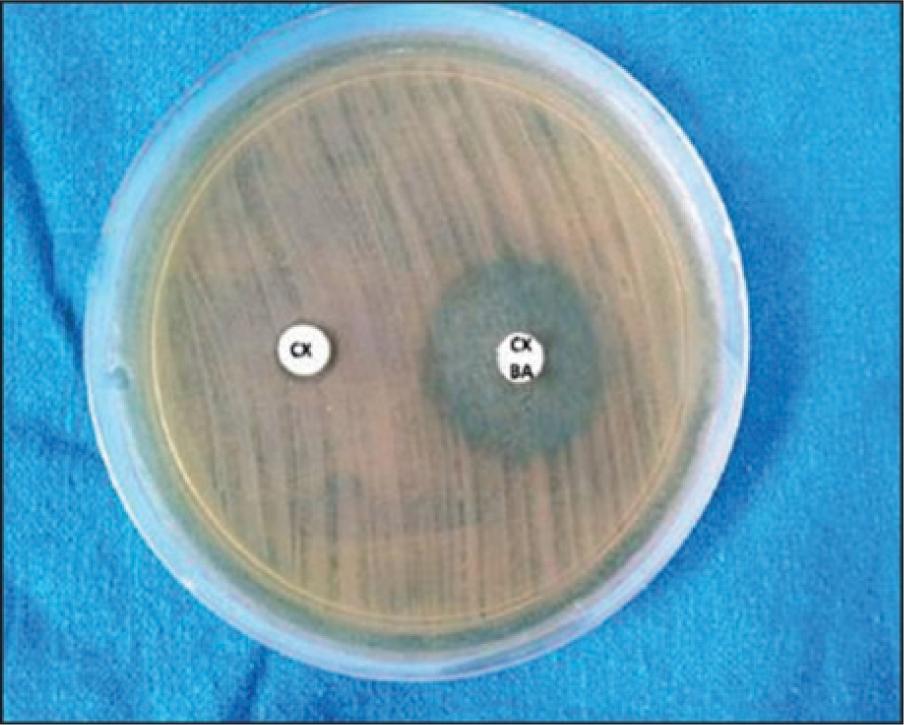
Conjugation frequency and cotransferred resistance markers
| Isolate No. | Protocol number | Frequency | Cotransferred resistance markers |
|---|---|---|---|
| 1 | 31 | 8×10-5 | Gm, Smx |
| 2 | 32 | 5.5×10-4 | |
| 3 | 35 | 3.5×10-5 | |
| 4 | 36 | 1.2×10-4 | |
| 5 | 37 | 1.4×10-5 | |
| 6 | 39 | 8.4×10-5 | |
| 7 | 40 | 4.5×10-5 | |
| 8 | 41 | 0 | |
| 9 | 43 | 10-4 | Tet |
| 10 | 44 | 5×10-4 | Tet |
| 11 | 45 | 1.6×10-6 | Tet |
| 12 | 46 | 4.5×10-6 | Tet |
| 13 | 47 | 1.6×10-4 | Tet |
| 14 | 49 | 0 | |
| 15 | 50 | 0 | |
| 16 | 51 | 7.2×10-6 | Smx |
| 17 | 53 | 1.57×10-5 | |
| 18 | 54 | 0 | |
| 19 | 55 | 5.6×10-6 | |
| 20 | 56 | 0 | |
| 21 | 58 | 0 | |
| 22 | 59 | 0 | |
| 23 | 60 | 0 | |
| 24 | 64 | 1.5×10-4 | |
| 25 | 65 | 1.2×10-4 | |
| 26 | 66 | 3.1×10-5 | Tet |
| 27 | 69 | 0 |
Antibiotic minimum inhibitory concentrations, genes, plasmids, and genotyping of E_ coli isolates from poultry farms in Bosnia and Herzegovina
| No. | Protocol number | Minimum inhibitory concentrations (mg/L) and resistance breakpoint (≥) of antibiotics | Genes, plasmids, and genotypes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX >32 | AMC 128/16 | TZP >128/4 | CZ >4 | CXM >32 | CAZ >32 | CTX >4 | CRO >4 | FEP >32 | IMI >4 | MEM >4 | GM >16 | CIP >4 | ESBL | bla genes | PBI and ST | ||
| 1 | 31 | >128 | 8 | 32 | >128 | >128 | 64 | >128 | >128 | 64 | 0.5 | 0.25 | >128 | >128 | + | TEM, CTX-M-15 | FIA |
| 2 | 32 | >128 | 32 | 8 | >128 | 32 | 0.5 | 32 | 32 | 0.25 | 0.12 | 0.06 | 2 | 0.25 | + | ND | ND |
| 3 | 35 | >128 | 1 | 2 | >128 | >128 | 2 | >128 | >128 | 0.5 | 0.06 | 0.06 | 0.5 | 2 | + | CTX-M-1 cluster | FIA |
| 4 | 36 | >128 | 4 | 8 | >128 | >128 | 2 | 64 | 64 | 1 | 0.06 | 0.06 | 1 | 0.25 | + | CTX-M-1 cluster | FIB, ST 117 |
| 5 | 37 | >128 | 8 | 16 | >128 | >128 | 1 | >128 | 64 | 1 | 0.12 | 0.06 | 1 | 0.5 | + | ND | FIB |
| 6 | 39 | >128 | 4 | 8 | >128 | >128 | 2 | 64 | 64 | 0.5 | 0.12 | 0.12 | 2 | 0.12 | + | TEM, CTX-M-15 | ND |
| 7 | 40 | >128 | 8 | 4 | >128 | >128 | 0.5 | >128 | >128 | 0.5 | 0.06 | 0.06 | 1 | 0.25 | + | ND | ND |
| 8 | 41 | >128 | 1 | 4 | >128 | 32 | 1 | 64 | 32 | 1 | 0.25 | 0.12 | 2 | 0.25 | + | CTX-M-1 cluster | ND |
| 9 | 43 | >128 | 16 | 16 | >128 | >128 | 8 | >128 | >128 | 1 | 0.25 | 0.06 | 4 | 0.06 | + | ND | ND |
| 10 | 44 | >128 | 32 | 16 | >128 | >128 | 32 | >128 | >128 | 2 | 0.06 | 0.06 | 0.5 | 0.12 | + | ND | FIB |
| 11 | 45 | >128 | 16 | 16 | >128 | >128 | 1 | >128 | >128 | 0.5 | 0.5 | 0.06 | 2 | 0.12 | + | CTX-M-1 cluster | ND |
| 12 | 46 | >128 | 8 | 4 | >128 | >128 | 2 | >128 | 64 | 1 | 0.12 | 0.12 | 4 | 0.06 | + | TEM, CTX-M-1 cluster | ND |
| 13 | 47 | >128 | 8 | 8 | >128 | >128 | 16 | 64 | 64 | 1 | 0.06 | 0.06 | 1 | 0.25 | + | TEM, CTX-M-1 cluster | I1 |
| 14 | 49 | >128 | 4 | 16 | >128 | >128 | 4 | >128 | >128 | 16 | 0.25 | 0.06 | 0,5 | 16 | + | TEM, CTX-M-1 cluster | I1 |
| 15 | 50 | >128 | 8 | 16 | >128 | >128 | 8 | >128 | >128 | 16 | 0.25 | 0.12 | 2 | 64 | + | TEM, CTX-M-15 | I1, FIB |
| 16 | 51 | >128 | 2 | 4 | >128 | >128 | 0.5 | >128 | >128 | 0.5 | 0.12 | 0.12 | 2 | 0.25 | + | ND | FIB |
| 17 | 53 | >128 | 2 | 4 | >128 | >128 | 8 | >128 | >128 | 8 | 0.25 | 0.06 | 1 | 0.25 | + | TEM, CTX-M-15 | ND |
| 18 | 54 | >128 | 4 | 2 | >128 | >128 | 1 | >128 | 64 | 0.5 | 0.5 | 0.06 | 0.5 | 0.12 | + | TEM, CTX-M-15 | I1, FIB |
| 19 | 55 | >128 | 1 | 2 | >128 | >128 | 1 | >128 | >128 | 2 | 0.06 | 0.06 | 0.5 | 0.5 | + | TEM, CTX-M-1 cluster | I1 |
| 20 | 56 | >128 | 4 | 32 | >128 | >128 | 16 | >128 | >128 | 16 | 0.5 | 0.25 | 2 | 4 | + | TEM, CTX-M-1 cluster | I1, HI1 |
| 21 | 58 | >128 | 2 | 2 | >128 | >128 | 2 | >128 | >128 | 8 | 0.25 | 0.25 | 1 | 0.25 | + | CTX-M-1 cluster | ND |
| 22 | 59 | >128 | 32 | 4 | >128 | 64 | 0.5 | >128 | 64 | 0.5 | 0.12 | 0.12 | 16 | 32 | + | TEM, CTX-M-1 cluster | I1 |
| 23 | 60 | >128 | 32 | 16 | >128 | >128 | 16 | >128 | 32 | 2 | 0.5 | 0.25 | 1 | 32 | + | CTX-M-1 cluster | I1, HI1, ST155 |
| 24 | 64 | >128 | 32 | 64 | >128 | 16 | >128 | >128 | 32 | 4 | 1 | 0.5 | 2 | >128 | + | TEM, CTX-M,-1 cluster | I1, FIB |
| 25 | 65 | >128 | 4 | 8 | >128 | >128 | 2 | 64 | 32 | 2 | 0.06 | 0.06 | 0.25 | 0.12 | + | TEM, CTX-M-1 cluster | I1 |
| 26 | 66 | >128 | 32 | 4 | >128 | 32 | >128 | >128 | 16 | 1 | 0.06 | 0.06 | 0.5 | 64 | + | TEM, CTX-M-1 cluster | FIB |
| 27 | 69 | >128 | 8 | 2 | >128 | >128 | 2 | >128 | >128 | 2 | 0.25 | 0.12 | 0.25 | 0.12 | + | CTX-M-15 | FIA |
Primers for ESBL detection with PCR
| Primer designation | Target gene | Sequence | Amplicon size | Ref. |
|---|---|---|---|---|
| OT 3 | TEM | 5’-ATG-AGT-ATT- CAA-CAT-TTC-CG-3’ | 850 | 25 |
| OT 4 | TEM | 5’-CCA-ATG-CTT-AAT-CAG-TGA-GG-3’ | 850 | 25 |
| SHV-F | SHV | 5’-TTC-GCC-TGT-GTA-TTA-TCT-CCC-3 | 1000 | 26 |
| SHV-R | SHV | 5’-TTA-GCG-TTG-CCA-GTG-YTC-GAT-3’ | 1000 | 26 |
| MA-1 | CTX-M | 5’-SCS-ATG-TGC-AGY-ACC-AGT-AA-3’ | 550 | 27 |
| MA-2 | CTX-M | 5’-CGC-CRA-TAT-GRT-TGG-TGG-TG-3’ | 550 | 27 |
| M1-F | CTX-M-1 cluster | 5’-AAA-AAT-CAC-TGC-GCC-AGT--TC-3’ | 415 | 30 |
| M1-R | CTX-M-1 cluster | 5’-TTG-GTG-ACG-ATT-TTA-GCC-GC-3’ | 415 | 30 |
| M2-F | CTX-M-2 cluster | 5’-CGA-CGC-TAC-CCC-TGC-TAT-T--3’ | 552 | 30 |
| M2-R | CTX-M-2 cluster | 5’-CCA-GCG-TCA-GAT-TTT-TCA-GG-3’ | 552 | 30 |
| M8-F | CTX-M-8 cluster | 5-TCG-CGT-TAA-GCG-GAT-GAT-GC | 666 | 30 |
| M9-F | CTX-M-9 cluster | 5’-CAA-AGA-GAG-TGC-AAC-GGA-TG | 205 | 30 |
| M9-R | CTX-M-9 cluster | 5’ATT-GGA-AAG-CGT-TCA-TCA-CC | 205 | 30 |
| M25-F | CTX-M-25 cluster | 5’GCA-CGA-TGA-CAT-TCG-GG | 327 | 30 |
| M9/M25-R | CTX-M-8/25 clusters | 5’AAC-CCA-CGA-TGT-GGG-TAG-C | 30 | |
| IS26-F | IS26 | 5’-AAA-AAT-GAT-TGA-AAG-GTG-GT-3’ | 31 | |
| IS26-R | IS26 | 5’-ATT-CGG-CAA-GTT-TTT-GCT-GT-3 | 31 | |
| ISEcp-F | ISEcp | 5’-AAA-AAT-GAT-TGA-AAG-GTG-GT-3’ | 31 | |
| ISEcp-R | ISEcp | 5’-AAT-ACT-ACC-TTG-CTT-TCT-GA-3’ | 31 | |
| QNR A-F | QNR A | 5’-ATT-TCT-CAC-GCC-AGG-ATT-TG-3’ | 29 | |
| QNR A-R | QNR A | 5’-GAT-CGG-CAA-AGG-TTA-GGT-CA-3’ | 29 | |
| QNR B-F | QNR B | 5’-GAT-CGT-GAA-AGC-CAG-AAA-GG | 29 | |
| QNR B-R | QNR B | 5’-ACG-ATG-CCT-GGT-AGT-TGT-CC | 29 | |
| QNR S-F | QNR S | 5’-ACG-ACA-TTC-GTC-AAC-TGC-AA | 29 | |
| QNR S-R | QNR S | 5’-TAA-ATT-GGC-ACC-CTG-TAG-GC | 29 |
Antibiotic susceptibility of E_ coli isolates established with the disk-diffusion test
| Isolate No. | Protocol number | Disk-diffusion method | |||
|---|---|---|---|---|---|
| FOX | AMI | ERT | SXT | ||
| 1 | 31 | S | S | S | R |
| 2 | 32 | S | S | S | S |
| 3 | 35 | S | S | S | S |
| 4 | 36 | S | S | S | S |
| 5 | 37 | S | S | S | S |
| 6 | 39 | S | S | S | S |
| 7 | 40 | S | S | S | S |
| 8 | 41 | S | S | S | S |
| 9 | 43 | S | S | S | S |
| 10 | 44 | S | S | S | S |
| 11 | 45 | S | S | S | S |
| 12 | 46 | S | S | S | S |
| 13 | 47 | S | S | S | S |
| 14 | 49 | S | S | S | S |
| 15 | 50 | S | S | S | S |
| 16 | 51 | S | S | S | S |
| 17 | 53 | S | S | S | S |
| 18 | 54 | S | S | S | S |
| 19 | 55 | S | S | S | S |
| 20 | 56 | S | S | S | R |
| 21 | 58 | R | S | S | S |
| 22 | 59 | S | S | S | S |
| 23 | 60 | R | S | S | S |
| 24 | 64 | R | S | S | S |
| 25 | 65 | S | S | S | S |
| 26 | 66 | R | S | S | S |
| 27 | 69 | S | S | S | S |