Fig. 1.

Fig. 2.
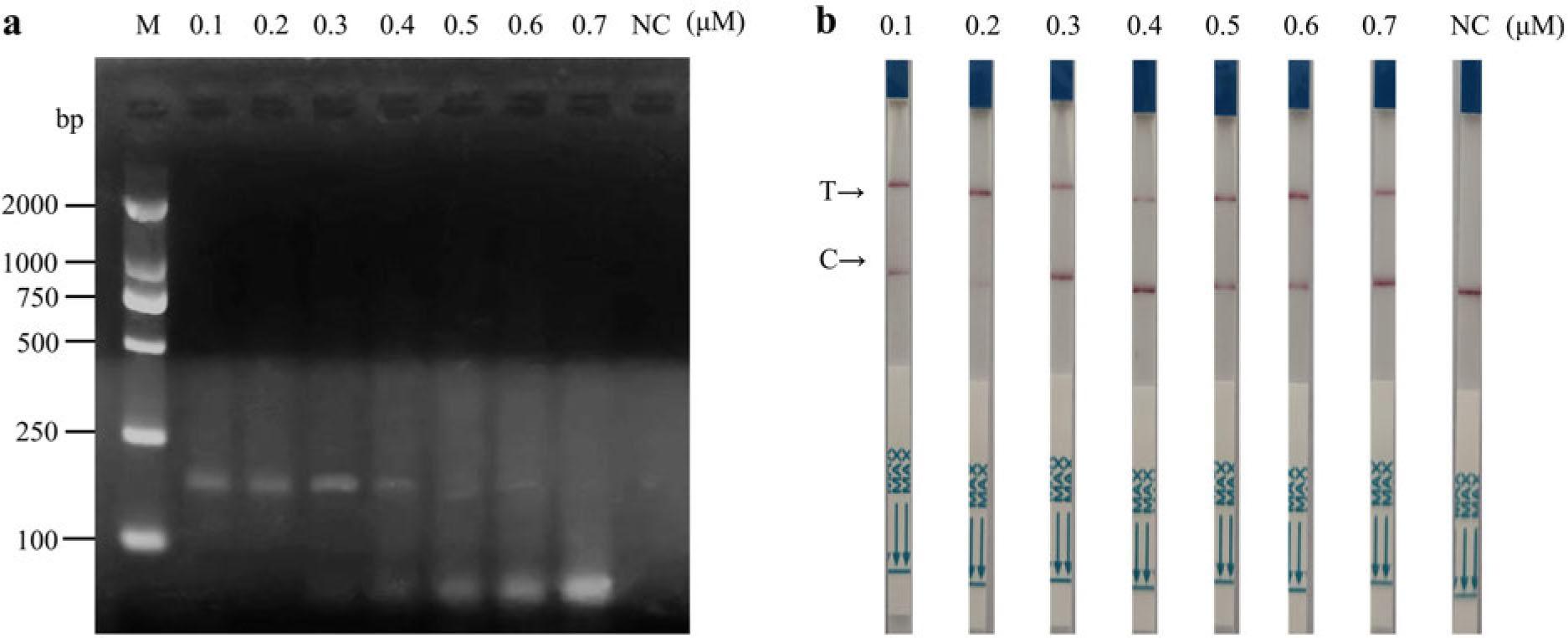
Fig. 3.
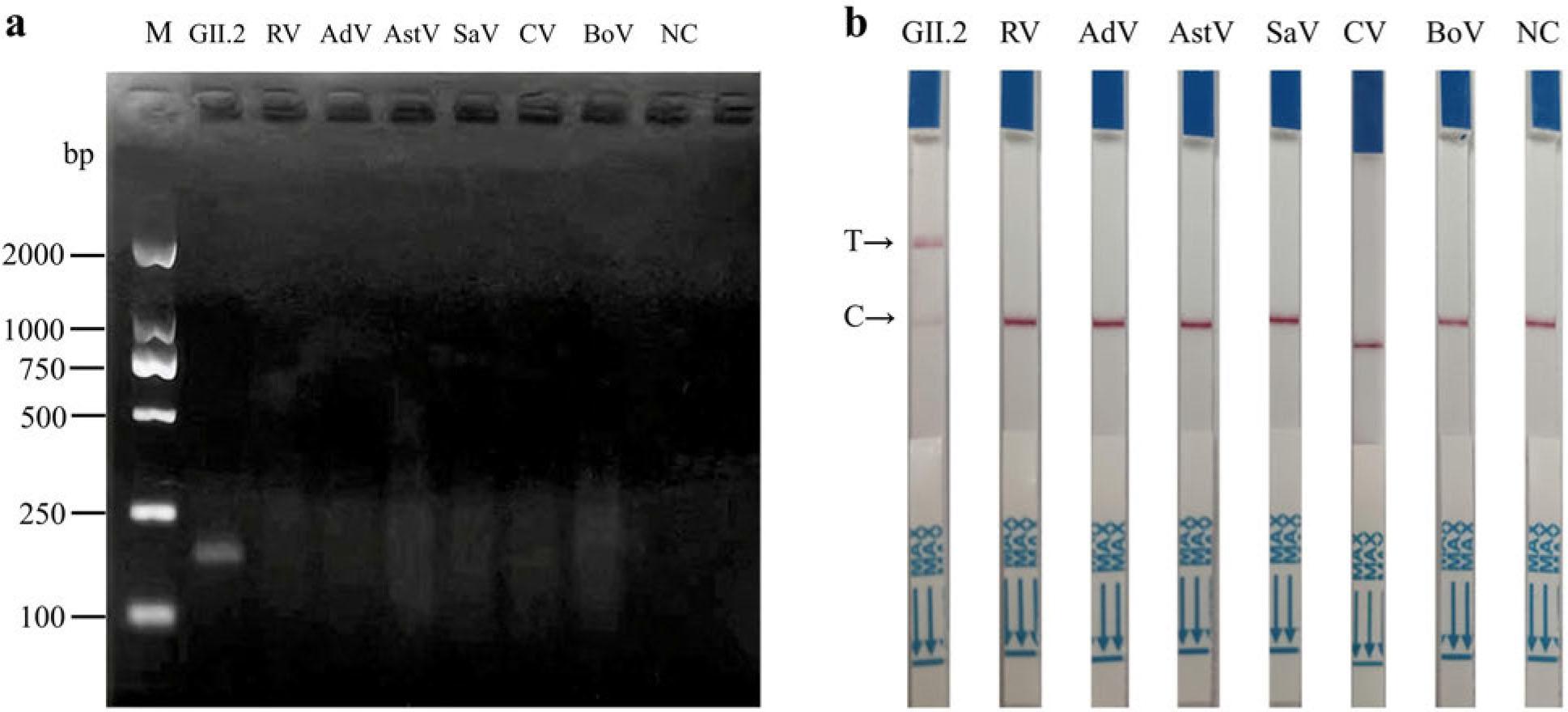
Fig. 4.
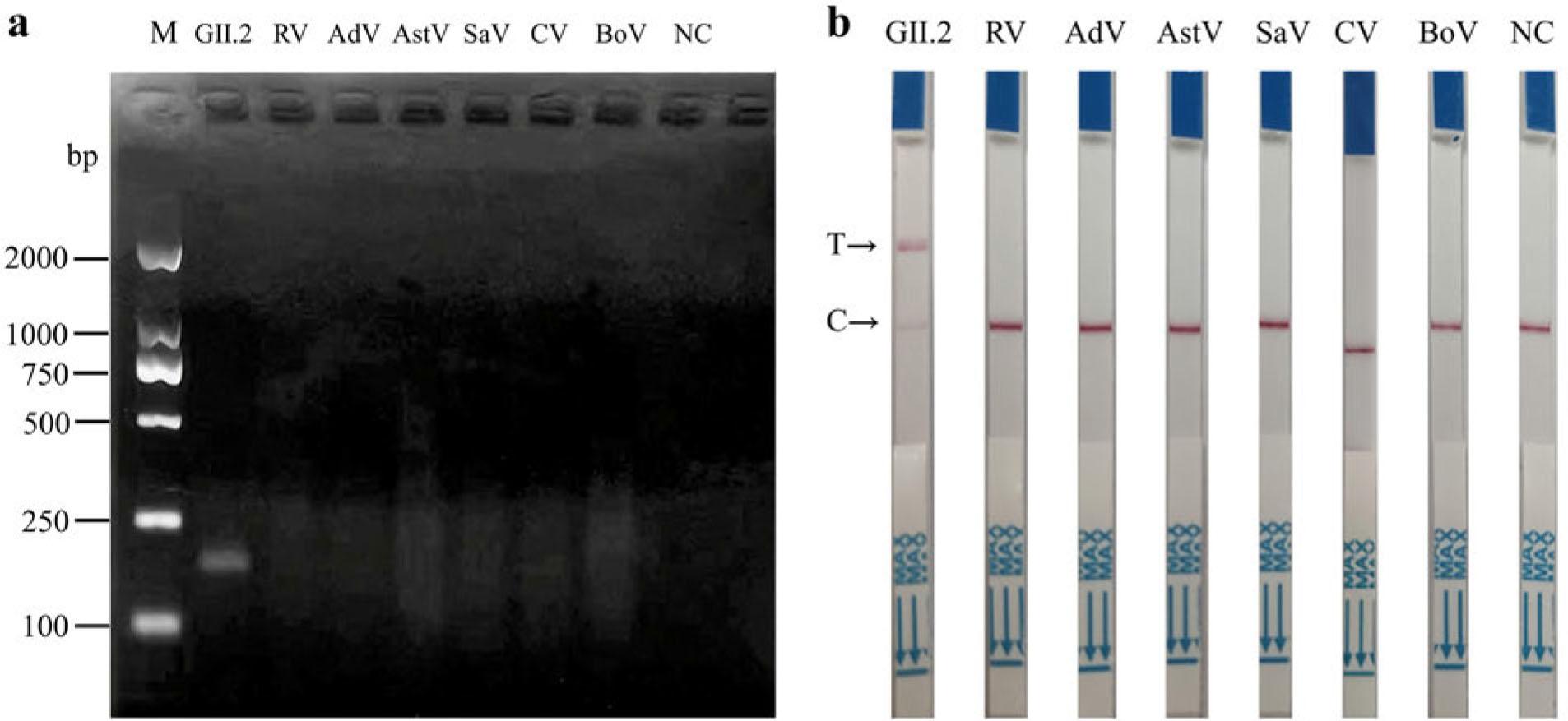
Fig. 5.
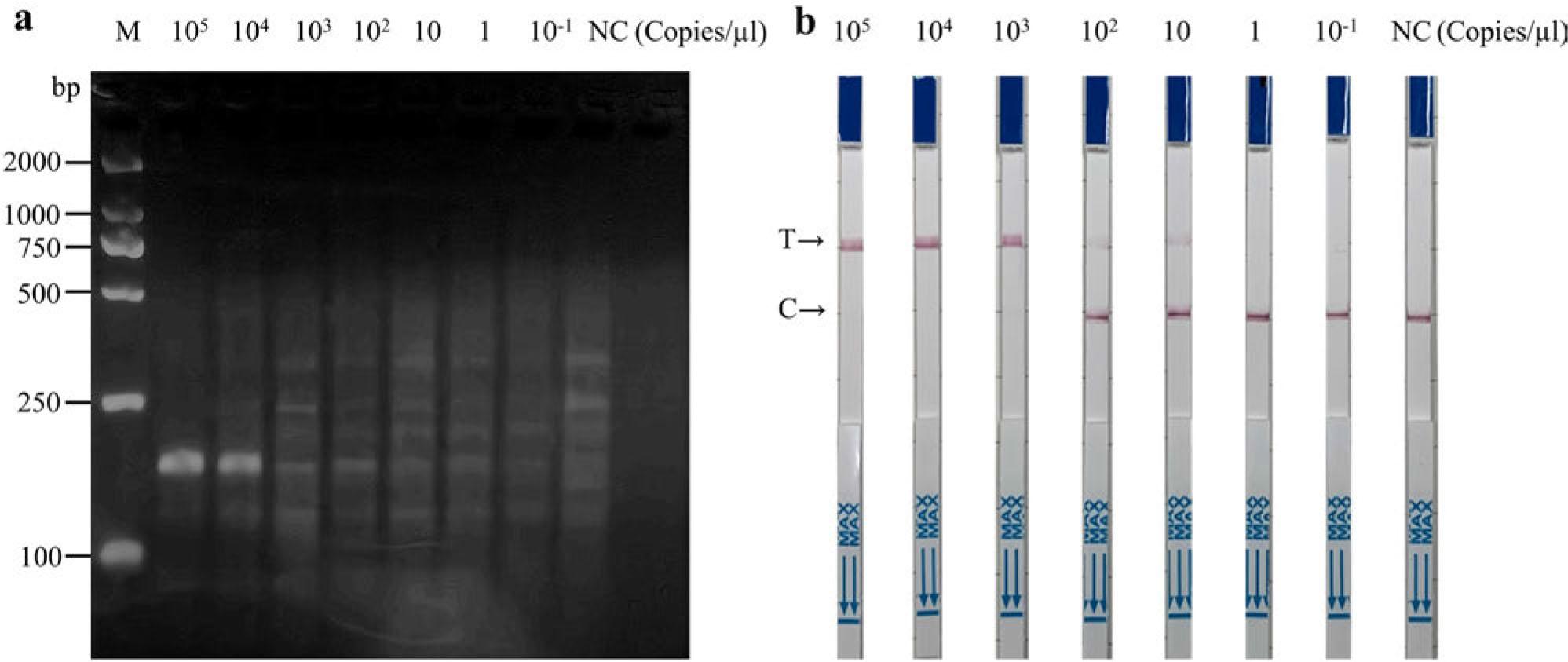
Fig. 6.

Fig. 7.
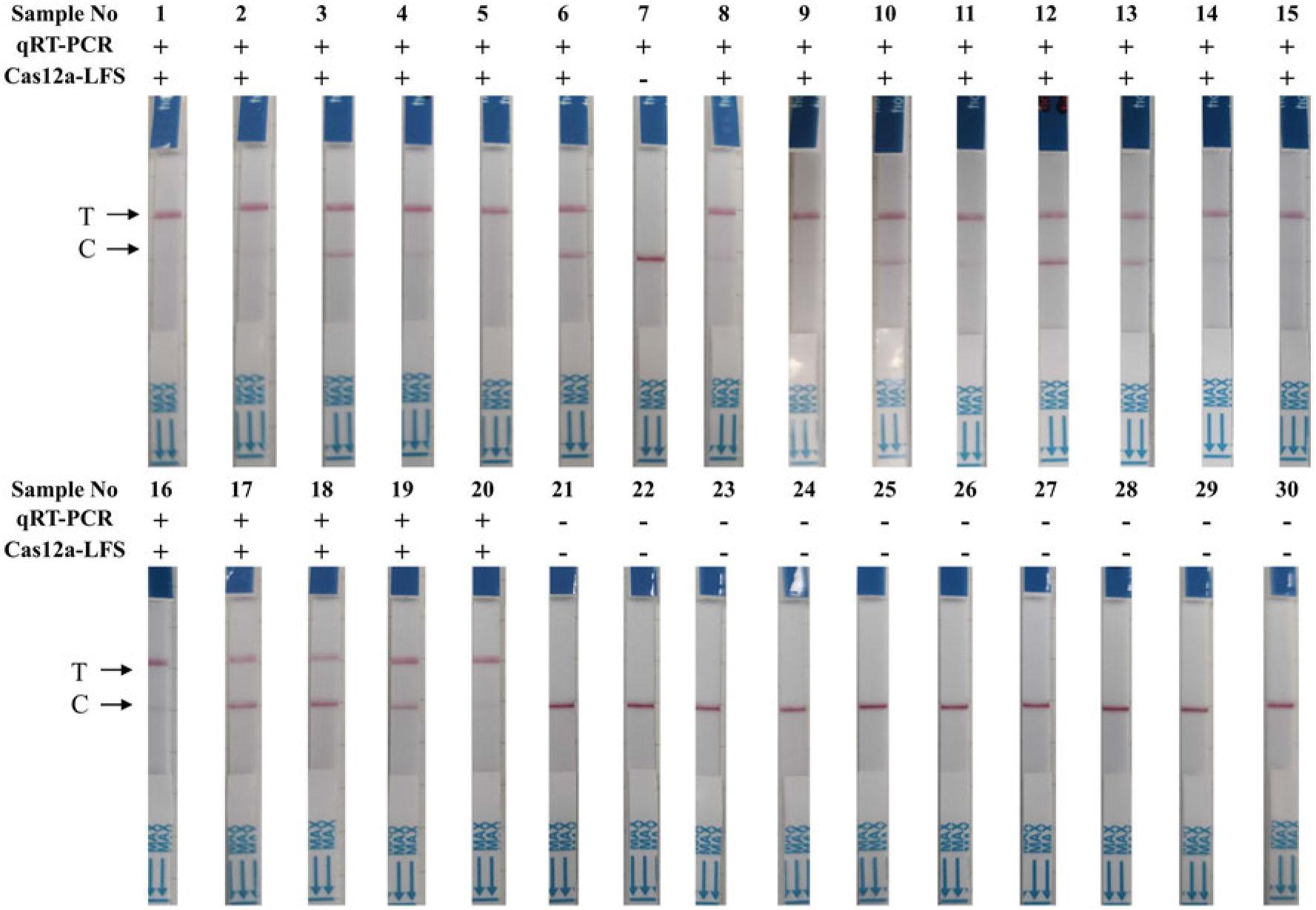
Statistical analysis of simulated samples using both the RT-RPA-Cas12a-LFS and qRT-PCR methods_
| qRT-PCR | CR | ||||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| RT-RPA-Cas12a-LFS | Positive | 38 | 0 | 38 | 98.33% |
| Negative | 1 | 21 | 22 | ||
| Total | 39 | 21 | 60 | ||
The oligonucleotide sequences for the primer, crRNA, and ssDNA reporter_
| Name Sequence (5’-3’) | NoV-GII.2-F1 AGGTGCYAATGCAATAAATCAGAGGGCAGA |
|---|---|
| NoV-GII.2-F2 | CAATGGGCTYAGTTCAYTRATYAATGCAGG |
| NoV-GII.2-R1 | CACCTCTGGCTGCATCAGCRGGGGAAAAGC |
| NoV-GII.2-R2 | TGTTTTATAGCCATCATRTCTGCCTGCAGC |
| Pre-NoV-GII.2-crRNA-F | GAAATTAATACGACTCACTATAGGGTAATTTCTACTAAGTGTAGATAATCATGATAAGGAGATGTT |
| Pre-NoV-GII.2-crRNA-R | AACATCTCCTTATCATGATTATCTACACTTAGTAGAAATTACCCTATAGTGAGTCGTATTAATTTC |
| ssDNA reporter | 6-FAM-TTATTATT-Biotin |
| NoV-GII.2-qPCR-F | GAGGGCAGAATTTGATTTTAATC |
| NoV-GII.2-qPCR-R | CCTTGTTTTATAGCCATCATG |
| NoV-GII.2-qPCR-P | FAM-TTGCCTGAATCTGAGCCTGC-BHQ1 |