Fig. 1

Fig. 2
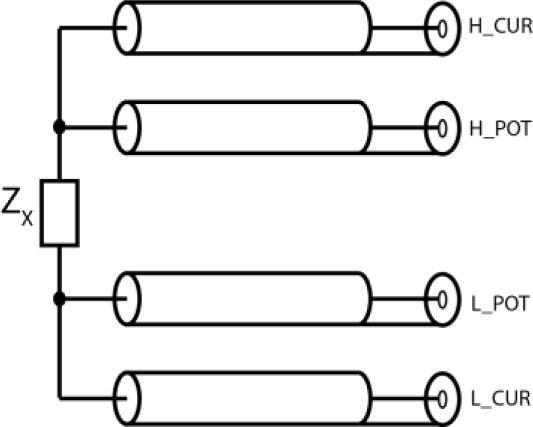
Fig. 3
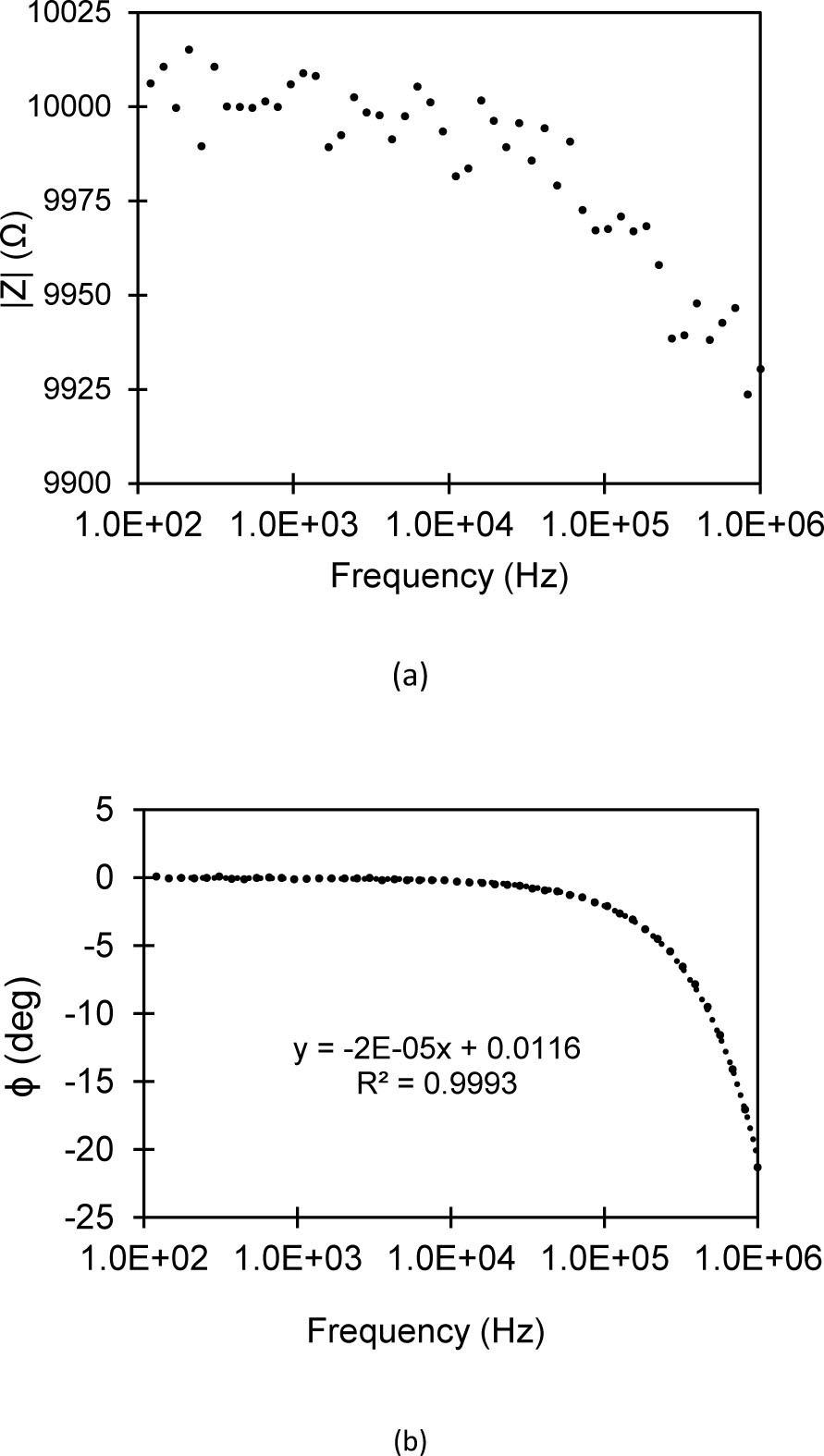
Fig. 4
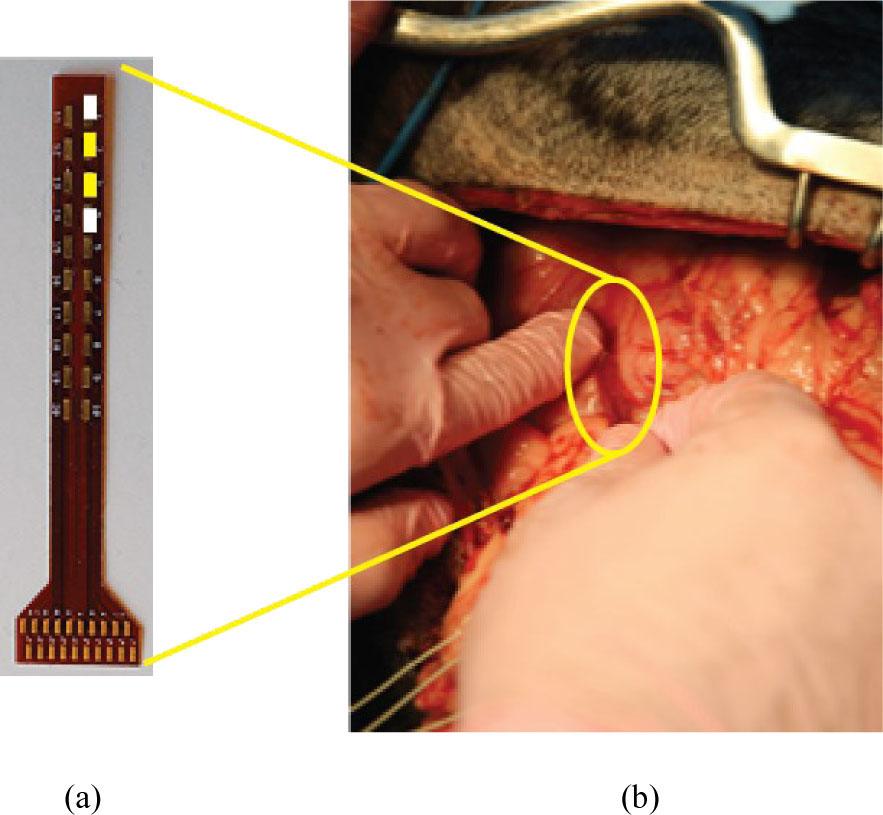
Fig. 5
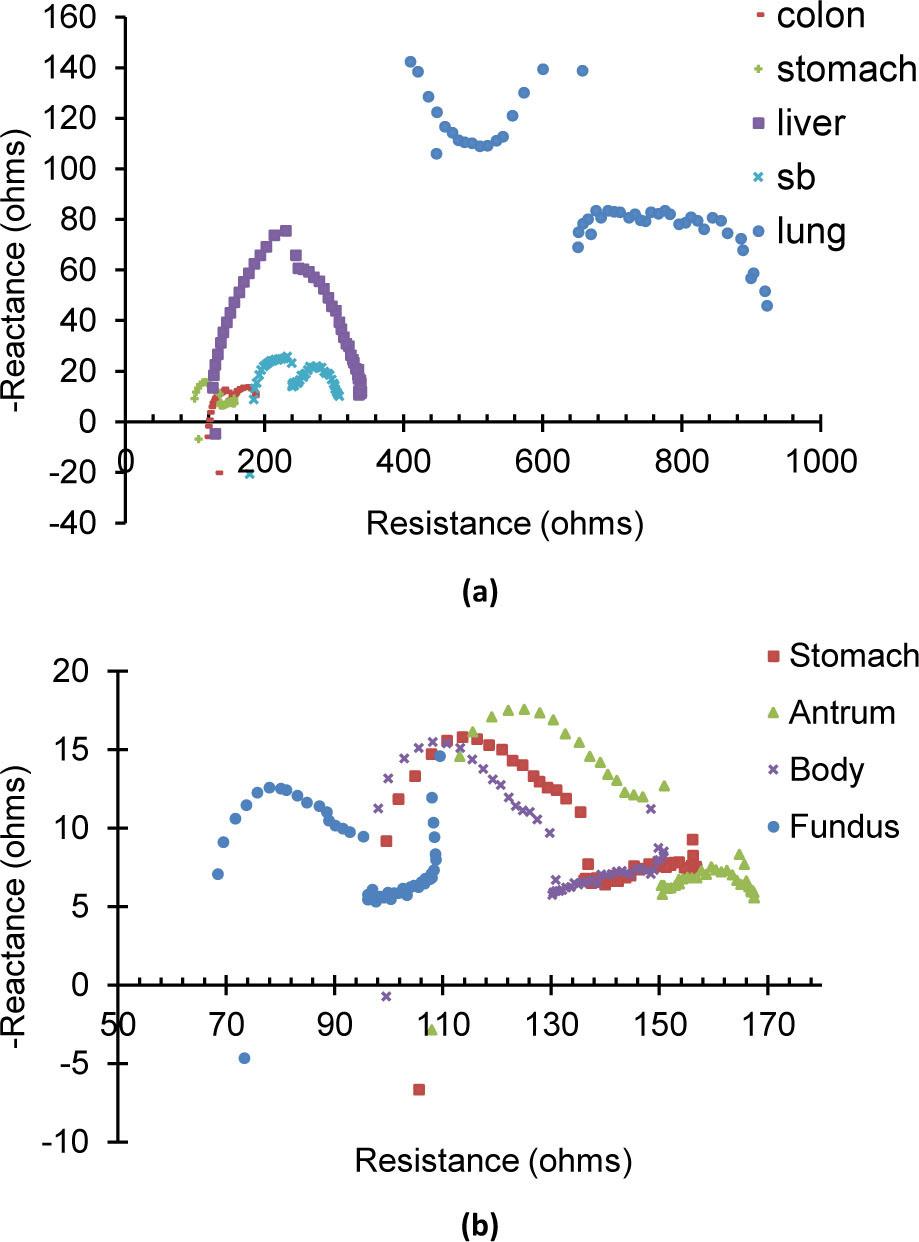
Fig. 6
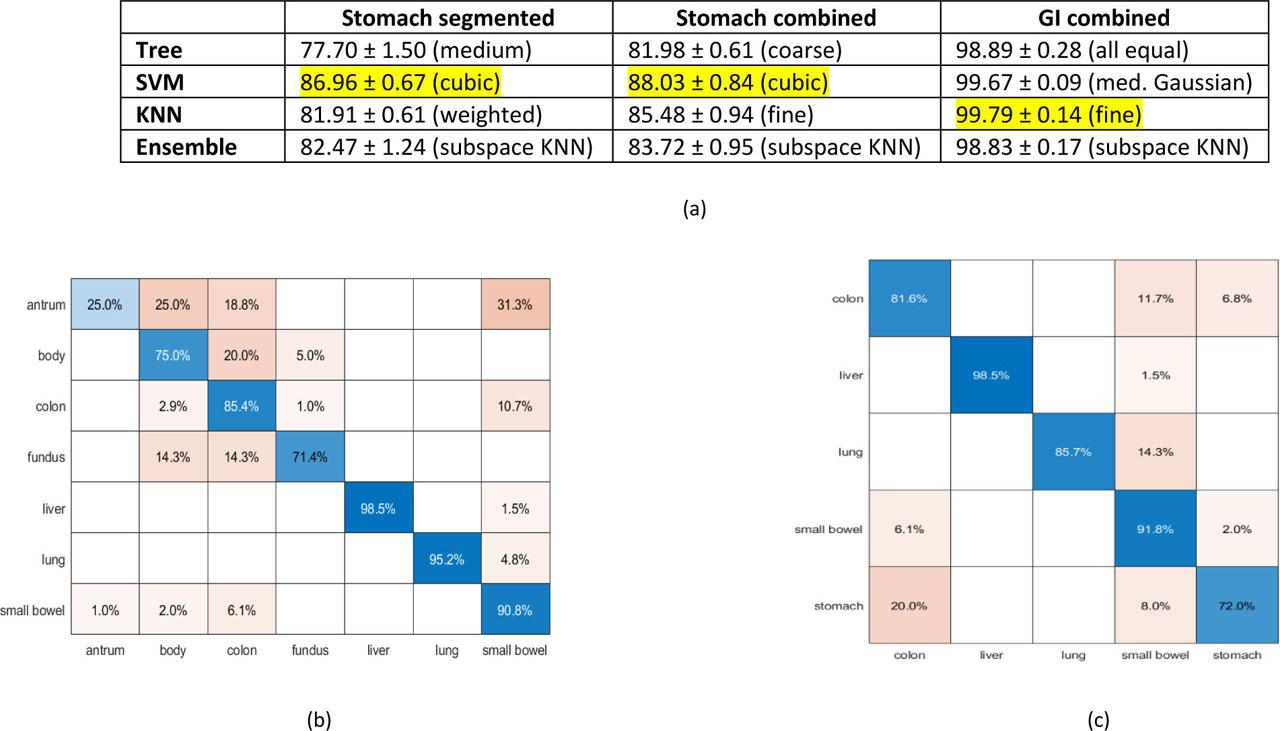






© 2021 Stephen Chiang, Matthew Eschbach, Robert Knapp, Brian Holden, Andrew Miesse, Steven Schwaitzberg, Albert Titus, published by University of Oslo
This work is licensed under the Creative Commons Attribution 4.0 License.