Figure 1.
![ANFIS network designed for the SCC strength modeling [28]](https://sciendo-parsed.s3.eu-central-1.amazonaws.com/6780fa99082aa65dea3c7504/j_acee-2024-0014_fig_001.jpg?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Content-Sha256=UNSIGNED-PAYLOAD&X-Amz-Credential=AKIA6AP2G7AKOUXAVR44%2F20251206%2Feu-central-1%2Fs3%2Faws4_request&X-Amz-Date=20251206T071222Z&X-Amz-Expires=3600&X-Amz-Signature=694d74b680fb10e47962f4443cfff480c2faf3e80482e8c42c97167067a85291&X-Amz-SignedHeaders=host&x-amz-checksum-mode=ENABLED&x-id=GetObject)
Figure 2.
![Hyperplanes of SVR [30]](https://sciendo-parsed.s3.eu-central-1.amazonaws.com/6780fa99082aa65dea3c7504/j_acee-2024-0014_fig_002.jpg?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Content-Sha256=UNSIGNED-PAYLOAD&X-Amz-Credential=AKIA6AP2G7AKOUXAVR44%2F20251206%2Feu-central-1%2Fs3%2Faws4_request&X-Amz-Date=20251206T071222Z&X-Amz-Expires=3600&X-Amz-Signature=9cbe5411ee4735bd8b60a94eaf39e9805eaced7be2db0719a626d0c1cbd2ad2c&X-Amz-SignedHeaders=host&x-amz-checksum-mode=ENABLED&x-id=GetObject)
Figure 3.
![ANN architecture [23]](https://sciendo-parsed.s3.eu-central-1.amazonaws.com/6780fa99082aa65dea3c7504/j_acee-2024-0014_fig_003.jpg?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Content-Sha256=UNSIGNED-PAYLOAD&X-Amz-Credential=AKIA6AP2G7AKOUXAVR44%2F20251206%2Feu-central-1%2Fs3%2Faws4_request&X-Amz-Date=20251206T071222Z&X-Amz-Expires=3600&X-Amz-Signature=c7044c0f4fd420311f4b240db77269bd5cc9ee225a80009b5911d5f651771230&X-Amz-SignedHeaders=host&x-amz-checksum-mode=ENABLED&x-id=GetObject)
Figure 4.
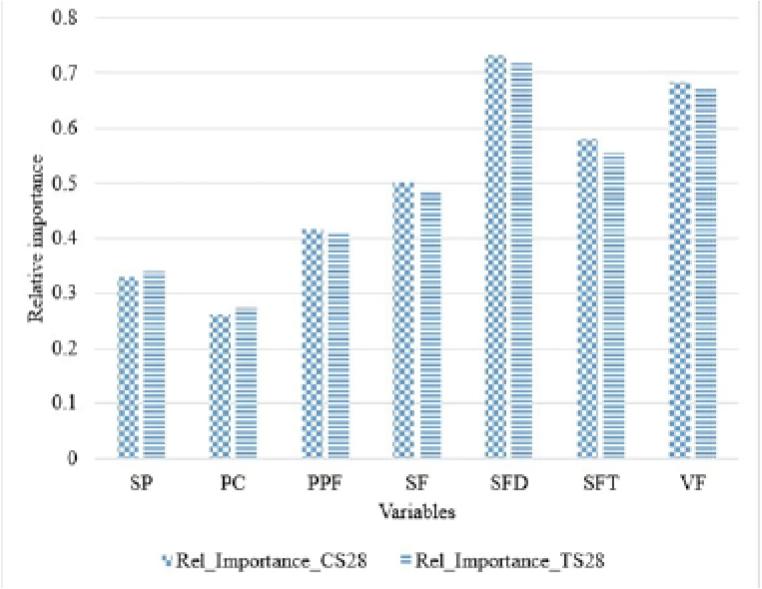
Figure 5.
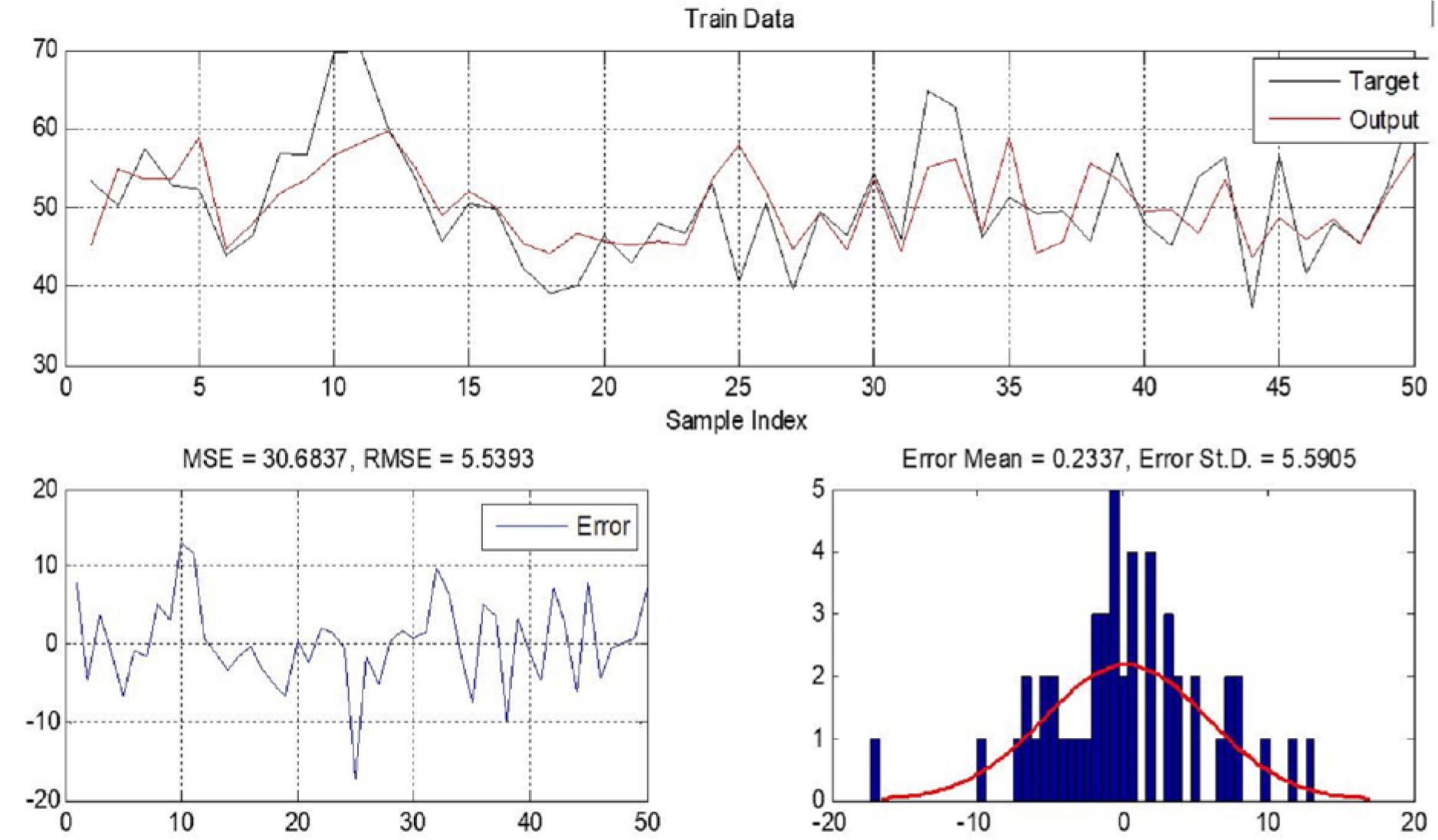
Figure 6.
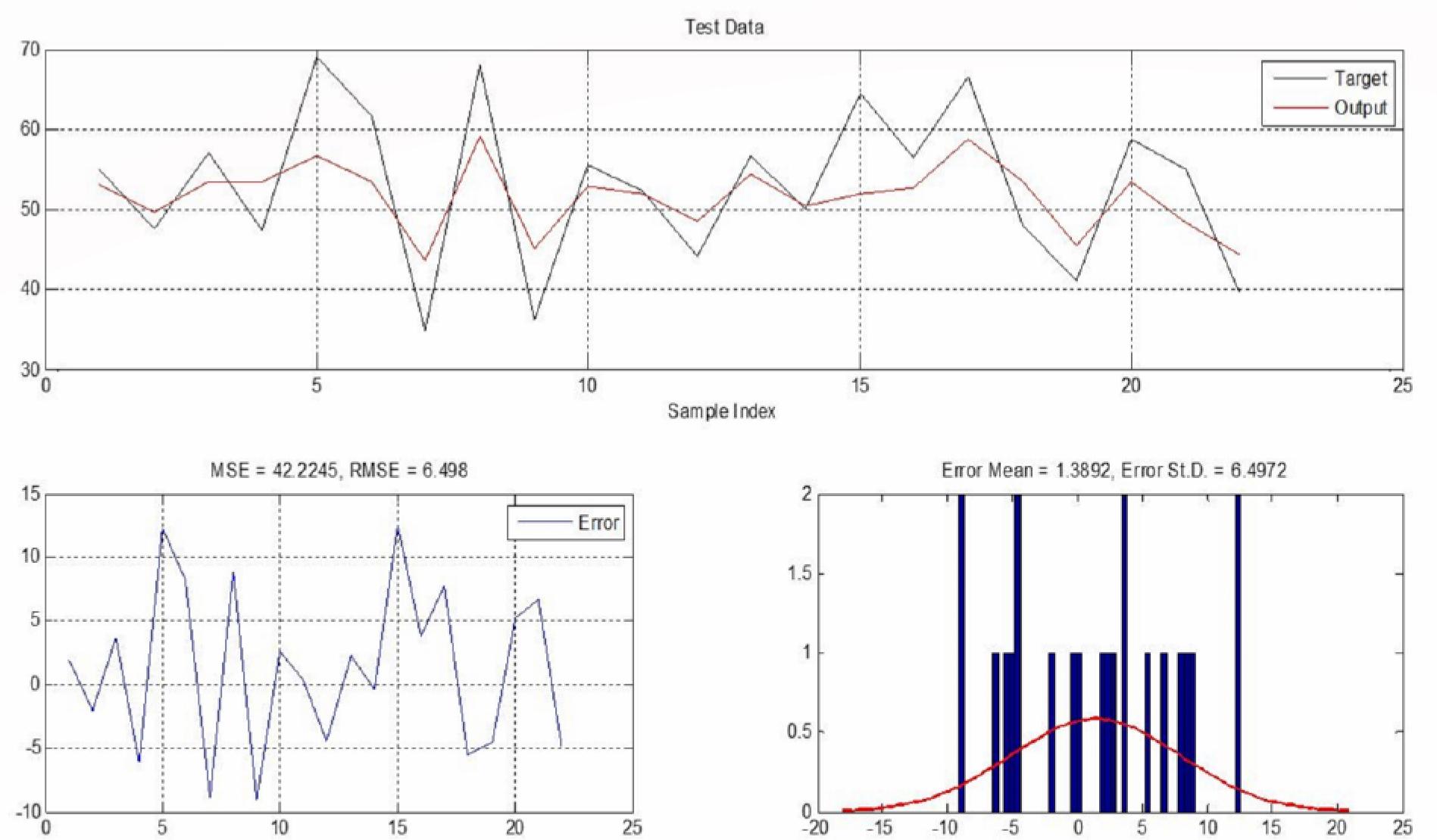
Figure 7.
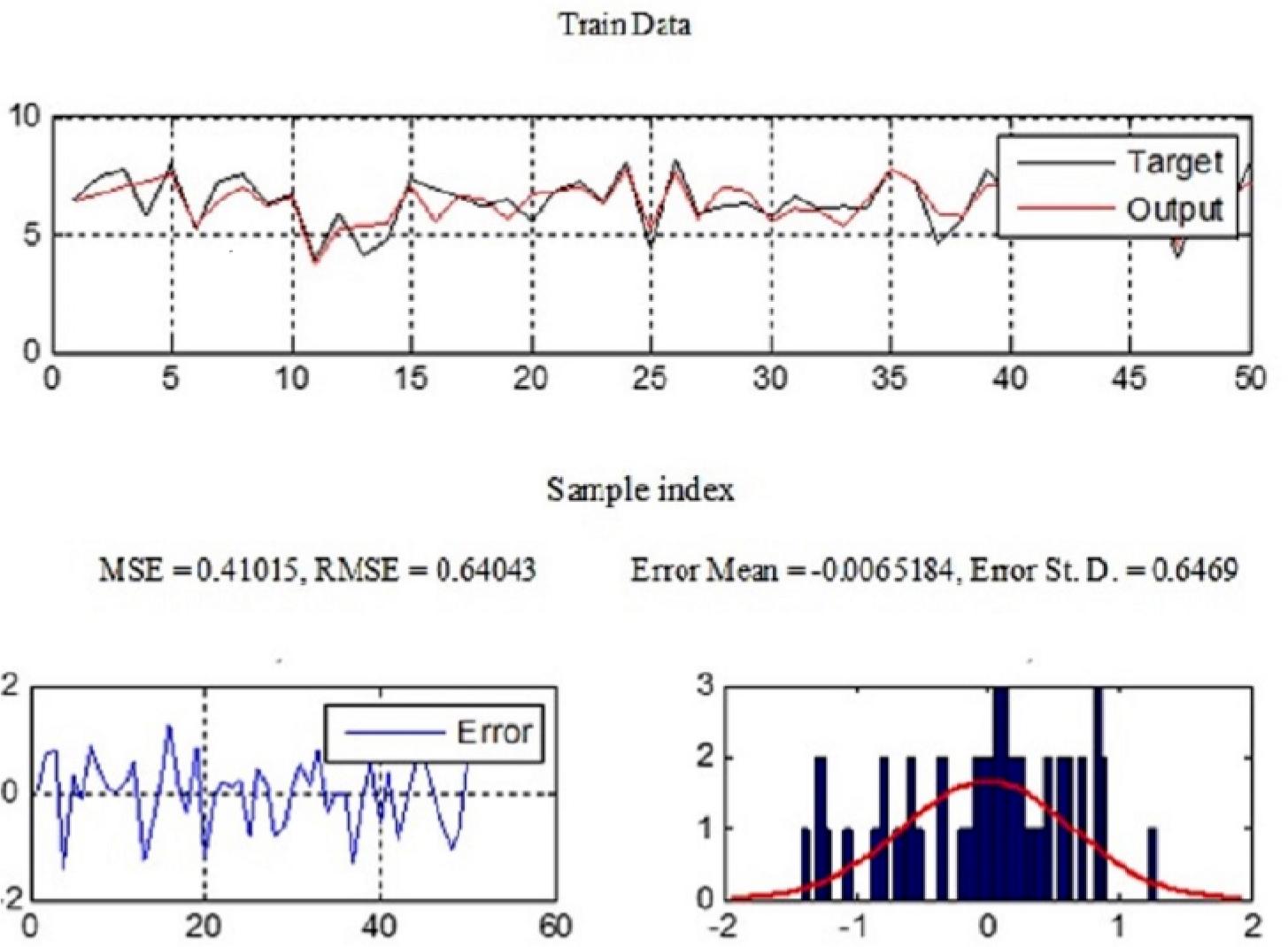
Figure 8.
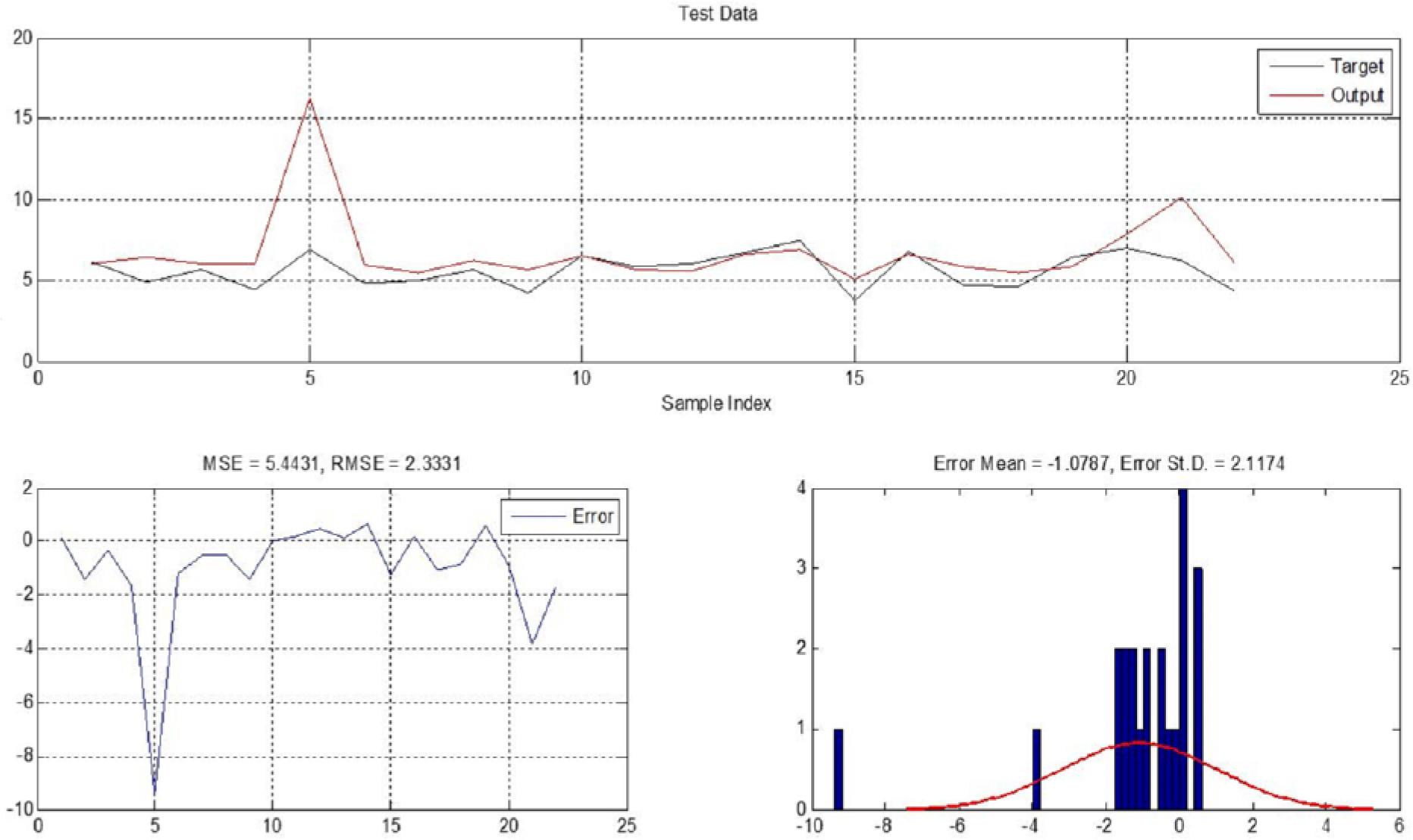
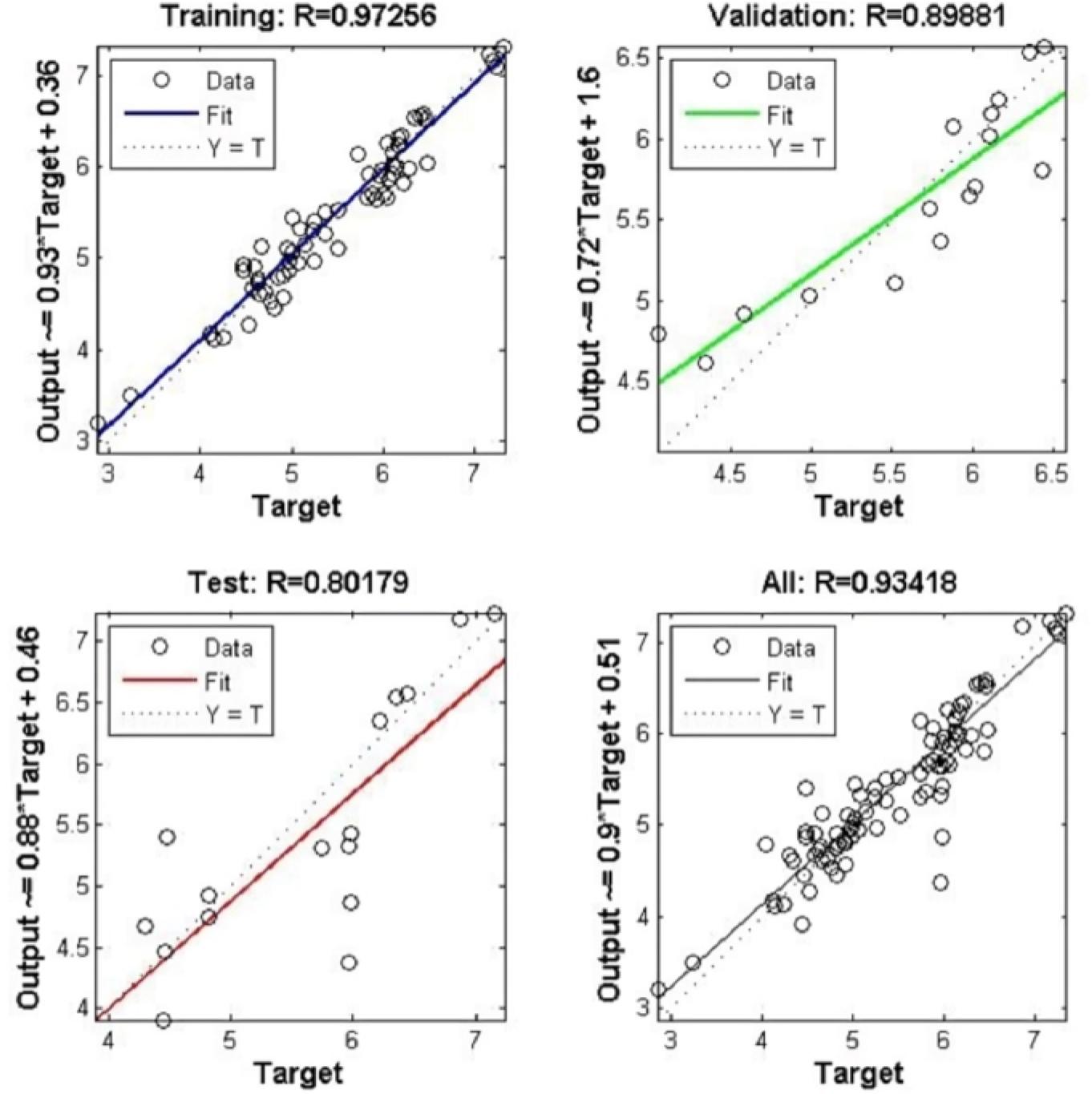
Figure 9.
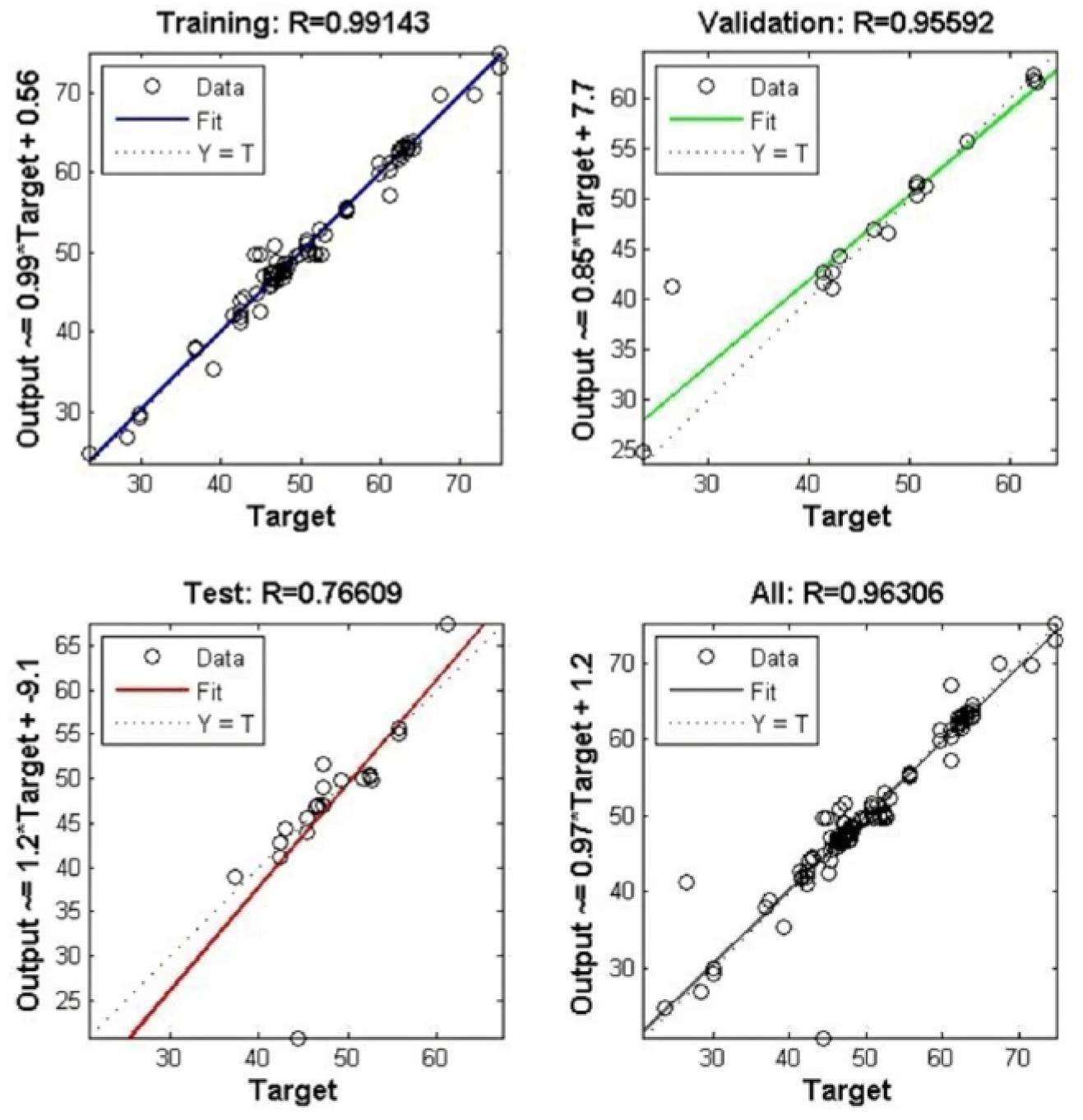
Figure 10.
SVM parameters used in the present study [20]
| SVM Parameter | Adoption/Values |
|---|---|
| Kernel function | RBF |
| Scaling factor | 1 |
| Method | Quadratic programming |
| Support Vectors | 13x5 |
| Bias | -0.012 |
Correlation matrix for TS28
| Variables | SP | PC | PPF | SF | SFD | SFT | VF | TS28 |
|---|---|---|---|---|---|---|---|---|
| SP | 1.00 | 0.14 | 0.00 | 0.00 | 0.42 | −0.36 | −0.32 | 0.71 |
| PC | 0.14 | 1.00 | 0.02 | 0.02 | 0.36 | −0.36 | −0.44 | 0.69 |
| PPF | 0.00 | 0.02 | 1.00 | −0.01 | −0.67 | 0.70 | 0.66 | 0.21 |
| SF | 0.00 | 0.02 | −0.01 | 1.00 | −0.30 | 0.23 | 0.31 | 0.29 |
| SFD | 0.42 | 0.36 | −0.67 | −0.30 | 1.00 | −0.98 | −0.96 | 0.26 |
| SFT | −0.36 | −0.36 | 0.70 | 0.23 | −0.98 | 1.00 | 0.97 | −0.23 |
| VF | −0.32 | −0.44 | 0.66 | 0.31 | −0.96 | 0.97 | 1.00 | −0.25 |
| TS28 | 0.71 | 0.69 | 0.21 | 0.29 | 0.26 | −0.23 | −0.25 | 1.00 |
Performance Matrix for CS28
| Models | Index of Agreement (IOA) | Akaike Information Criterion (AIC) | Skill Score (SS) | Symmetric Uncertainty (SU) |
|---|---|---|---|---|
| ANFIS | 0.50 | 256.34 | 0.50 | 0.01 |
| ANN | 0.64 | 232.34 | 0.64 | 0.29 |
| SVM | 0.96 | 68.33 | 0.96 | 0.93 |
| MLR | 0.51 | 256.04 | 0.51 | 0.01 |
| GEP | 0.62 | 159.31 | 0.58 | 0.73 |
Performance Matrix for TS28
| Models | Index of Agreement (IOA) | Akaike Information Criterion (AIC) | Skill Score (SS) | Symmetric Uncertainty (SU) |
|---|---|---|---|---|
| ANFIS | 0.67 | −59.96 | 0.67 | 0.33 |
| ANN | −0.54 | 50.01 | −0.54 | −2.07 |
| SVM | 0.81 | −99.25 | 0.81 | 0.61 |
| MLR | 0.87 | −127.64 | 0.87 | 0.74 |
| GEP | 0.62 | −51.32 | 0.62 | 0.25 |
Correlation matrix for CS28
| VARIABLES | SP | PC | PPF | SF | SFD | SFT | VF | CS28 |
|---|---|---|---|---|---|---|---|---|
| SP | 1.00 | 0.14 | 0.00 | 0.00 | 0.42 | −0.36 | −0.32 | 0.68 |
| PC | 0.14 | 1.00 | 0.02 | 0.02 | 0.36 | −0.36 | −0.44 | 0.72 |
| PPF | 0.00 | 0.02 | 1.00 | −0.01 | −0.67 | 0.70 | 0.66 | 0.16 |
| SF | 0.00 | 0.02 | −0.01 | 1.00 | −0.30 | 0.23 | 0.31 | 0.32 |
| SFD | 0.42 | 0.36 | −0.67 | −0.30 | 1.00 | −0.98 | −0.96 | 0.28 |
| SFT | −0.36 | −0.36 | 0.70 | 0.23 | −0.98 | 1.00 | 0.97 | −0.25 |
| VF | −0.32 | −0.44 | 0.66 | 0.31 | −0.96 | 0.97 | 1.00 | −0.26 |
| CS28 | 0.68 | 0.72 | 0.16 | 0.32 | 0.28 | −0.25 | −0.26 | 1.00 |
Initial values of ANFIS GA parameters [28]
| Parameters | Values |
|---|---|
| Population size | 20 |
| Iterations | 1000 |
| Crossover rate | 0.70 |
| Mutation rate | 0.50 |
| Inversion rate | 0.10 |
| Selection pressure | 8.0 |
| Gamma | 0.20 |
Dataset for training and testing of different models
| SP (kg) | PC (kg) | PPF (kg) | SF (kg) | SFD (Dia) | SFT (Sec) | VF (Sec) | CS28 (MPa) | TS28 (MPa) | Water (kg) | FA (kg) | CA1 20-10 mm (kg) | CA2 10-4.75 mm (kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 600 | 0 | 0 | 780 | 2 | 7 | 36.11 | 4.41 | 240 | 810 | 365 | 365 |
| 9 | 600 | 1.5 | 3 | 665 | 5 | 13 | 39.95 | 4.79 | 240 | 810 | 365 | 365 |
| 9 | 600 | 3 | 6 | 630 | 6 | 14 | 41.1 | 5.26 | 240 | 810 | 365 | 365 |
| 9.225 | 615 | 0 | 0 | 790 | 1.5 | 6 | 39.55 | 4.67 | 246 | 810 | 365 | 365 |
| 9.225 | 615 | 1.535 | 3.07 | 730 | 3 | 9 | 42.33 | 5.66 | 246 | 810 | 365 | 365 |
| 9.225 | 615 | 3.07 | 6.15 | 690 | 4 | 11 | 46.31 | 6.12 | 246 | 810 | 365 | 365 |
| 9.45 | 630 | 0 | 3.15 | 750 | 2.5 | 8 | 45.52 | 6.03 | 252 | 810 | 365 | 365 |
| 9.45 | 630 | 1.575 | 6.3 | 730 | 3 | 8 | 49.16 | 6.88 | 252 | 810 | 365 | 365 |
| 9.45 | 630 | 3.15 | 0 | 725 | 3 | 9 | 46.46 | 6.28 | 252 | 810 | 365 | 365 |
| 12 | 600 | 0 | 6 | 760 | 2 | 8 | 44.12 | 5.86 | 240 | 810 | 365 | 365 |
| 12 | 600 | 1.5 | 0 | 765 | 2 | 7 | 41.68 | 5.53 | 240 | 810 | 365 | 365 |
| 12 | 600 | 3 | 3 | 710 | 4 | 12 | 45.09 | 6.22 | 240 | 810 | 365 | 365 |
| 12.3 | 615 | 0 | 3.07 | 775 | 2 | 7 | 48.03 | 6.74 | 246 | 810 | 365 | 365 |
| 12.3 | 615 | 1.535 | 6.15 | 725 | 3 | 9 | 53.11 | 7.26 | 246 | 810 | 365 | 365 |
| 12.3 | 615 | 3.07 | 0 | 710 | 4 | 10 | 47.36 | 6.67 | 246 | 810 | 365 | 365 |
| 12.6 | 630 | 0 | 6.3 | 810 | 1 | 6 | 57.16 | 7.8 | 252 | 810 | 365 | 365 |
| 12.6 | 630 | 1.575 | 0 | 785 | 2 | 6 | 54.3 | 7.52 | 252 | 810 | 365 | 365 |
| 12.6 | 630 | 3.15 | 3.15 | 750 | 3 | 9 | 56.44 | 7.76 | 252 | 810 | 365 | 365 |
| 3.3 | 565 | 0 | 0 | 690 | 2 | 11 | 52.8 | 3.9 | 186 | 750 | 58.5 | 391.5 |
| 5 | 566.7 | 0 | 0 | 680 | 2.2 | 16 | 57.3 | 4 | 170 | 760 | 58.5 | 391.5 |
| 5.1 | 566.7 | 0 | 39 | 665 | 2.8 | 18 | 56.9 | 5.9 | 170 | 760 | 58.5 | 391.5 |
| 5.3 | 566.7 | 4.55 | 39 | 670 | 3.1 | 19 | 61.7 | 6.9 | 170 | 760 | 58.5 | 391.5 |
| 5.7 | 566.7 | 6.825 | 39 | 660 | 3.3 | 18 | 58.8 | 7.2 | 170 | 760 | 58.5 | 391.5 |
| 6.2 | 566.7 | 9.1 | 39 | 645 | 4.2 | 20 | 56.7 | 6.9 | 170 | 760 | 58.5 | 391.5 |
| 10 | 500 | 0 | 0 | 700 | 2.4 | 7 | 37.21 | 4.25 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 157 | 600 | 4 | 12 | 51.31 | 7.2 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 117.75 | 620 | 3.8 | 11 | 50.24 | 6.25 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 78.5 | 640 | 3.5 | 10.5 | 50.45 | 6.1 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 39.25 | 650 | 3.5 | 10.5 | 45.64 | 4.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 0 | 670 | 3 | 8.3 | 34.87 | 4.1 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 157 | 580 | 5 | 12.7 | 40.52 | 5.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 117.75 | 600 | 4 | 12 | 45.78 | 5.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 78.5 | 620 | 3.5 | 11 | 50.44 | 6.25 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 39.25 | 630 | 3.5 | 10.6 | 47.86 | 5.85 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 0 | 640 | 3.2 | 10.5 | 47.84 | 5.75 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 157 | 560 | 5.5 | 13 | 64.1 | 7.25 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 117.75 | 570 | 5.5 | 12.5 | 54.98 | 6.15 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 78.5 | 590 | 5 | 11.5 | 47.64 | 5.7 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 39.25 | 610 | 4.5 | 11 | 46.09 | 5.75 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 0 | 620 | 4 | 11 | 43.96 | 3.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 0 | 720 | 2.2 | 6.8 | 39.04 | 4.3 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 157 | 620 | 4 | 11.6 | 66.69 | 7.95 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 117.75 | 630 | 4 | 11 | 53.91 | 6.5 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 78.5 | 640 | 3.5 | 10.5 | 52.31 | 6.5 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 39.25 | 660 | 3.5 | 10.5 | 47.96 | 5.7 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 0 | 680 | 2.5 | 8.1 | 39.53 | 4.6 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 157 | 600 | 4.5 | 12.5 | 52.27 | 6.45 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 117.75 | 620 | 4 | 12 | 62.67 | 7.35 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 78.5 | 630 | 4 | 11 | 56.92 | 6.5 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 39.25 | 640 | 3.5 | 10.4 | 55.13 | 6.45 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 0 | 650 | 3.5 | 10.2 | 53.21 | 6.2 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 157 | 580 | 5.5 | 12.5 | 69.12 | 8.1 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 117.75 | 590 | 5.5 | 12 | 55.68 | 6.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 78.5 | 600 | 5 | 11.8 | 50.05 | 6.4 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 39.25 | 620 | 4 | 11 | 46.5 | 5.9 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 0 | 630 | 4 | 10.6 | 46.05 | 4.4 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 0 | 740 | 2 | 6.8 | 42.83 | 4.95 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 157 | 630 | 4 | 11.6 | 68.14 | 8.05 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 117.75 | 630 | 4 | 11 | 64.79 | 7.5 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 78.5 | 650 | 3.2 | 10.5 | 56.55 | 6.65 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 39.25 | 660 | 3 | 10.5 | 49.82 | 5.9 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0 | 0 | 700 | 2.5 | 8.1 | 46.72 | 4.75 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 157 | 620 | 4.5 | 12.5 | 60.22 | 7 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 117.75 | 630 | 4 | 12 | 69.71 | 8.15 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 78.5 | 640 | 4 | 11 | 64.52 | 7.4 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 39.25 | 650 | 3.5 | 10.4 | 56.7 | 6.6 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.455 | 0 | 670 | 3 | 10.2 | 53.73 | 6.55 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 157 | 600 | 5 | 12.5 | 69.79 | 8.15 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 117.75 | 610 | 5 | 12 | 56.67 | 7.75 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 78.5 | 620 | 4.5 | 11.8 | 52.48 | 6.8 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 39.25 | 640 | 3.5 | 11 | 49.6 | 6.15 | 190 | 1005 | 310 | 310 |
| 10 | 500 | 0.91 | 0 | 650 | 3.5 | 10.6 | 49.4 | 5 | 190 | 1005 | 310 | 310 |
GEP parameters [27]
| Parameters | Values |
|---|---|
| Population size | 30 |
| Genes per chromosome | 3 |
| Gene head length | 9 |
| Functions | +, -./, *,^ |
| Gene tail length | 12 |
| Mutation rate | 0.05 |
| Inversion rate | 0.1 |
| Gene transposition rate | 0.1 |
| One-point recombination rate | 0.3 |
| Two-point recombination rate | 0.3 |
| Gene recombination rate | 0.1 |
| Fitness function | R≥0.7 |