Figure S1:
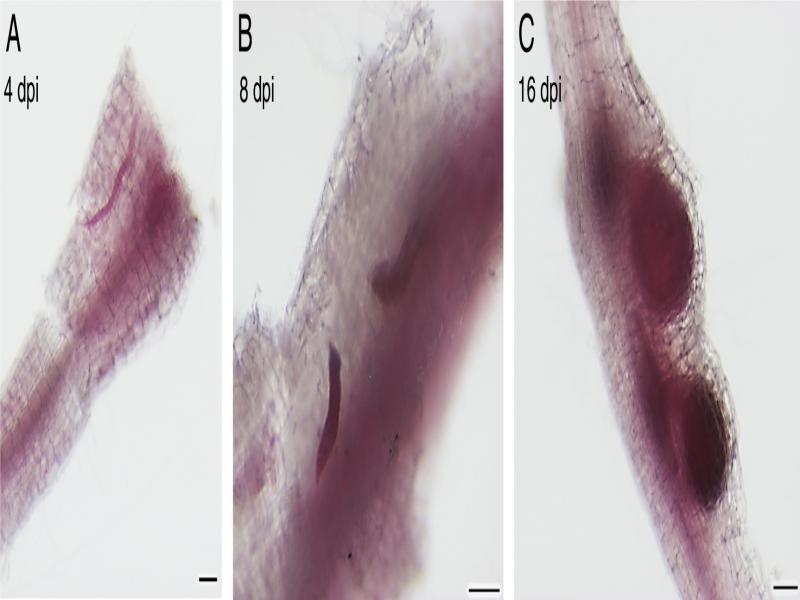
Figure 1:
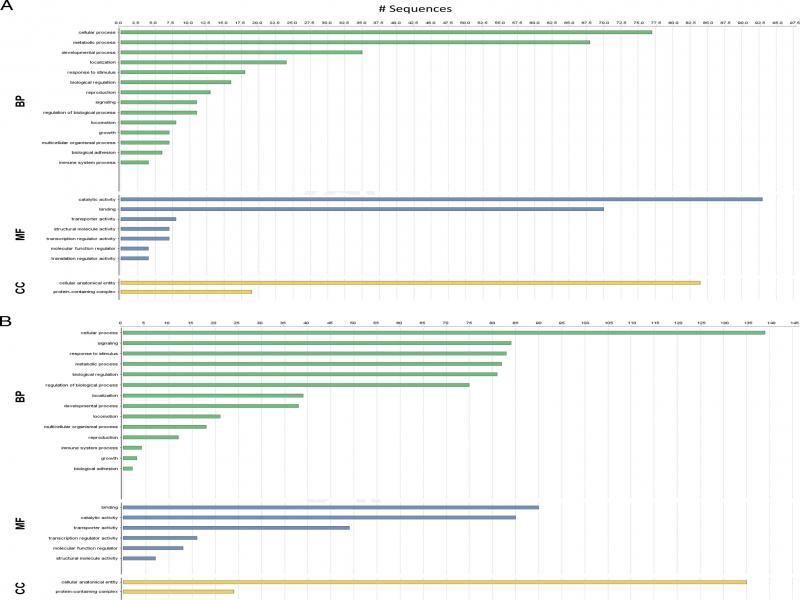
Figure 2:
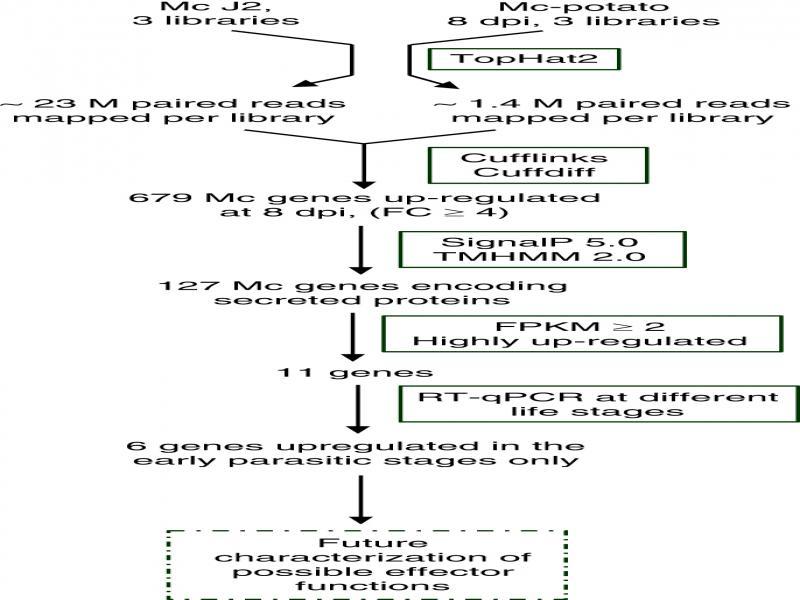
Figure 3:
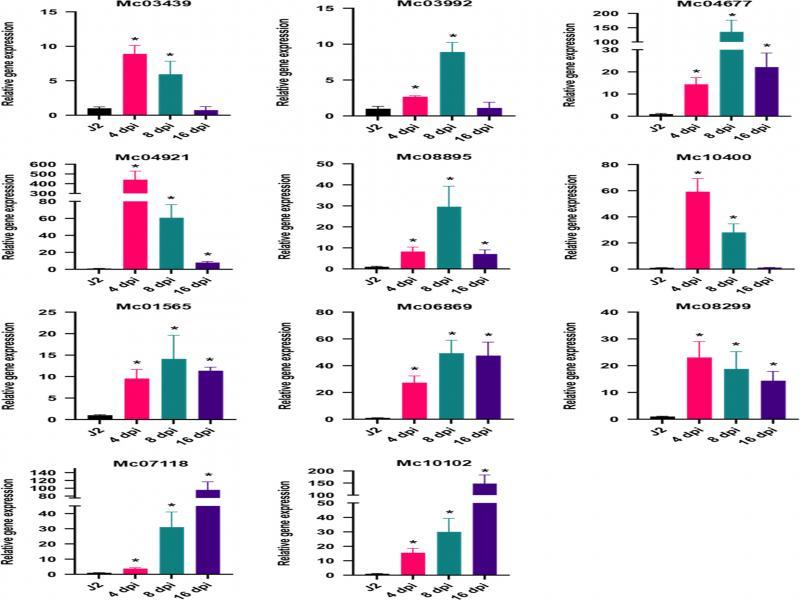
Figure 4:

Meloidogyne chitwoodi RNA-seq libraries and mapping to genome_
| Library | Bio-replicate | Read pairs | Read pairs mapped to genome | Mapping rate (%) |
|---|---|---|---|---|
| Pre-parasitic J2 | A | 51 950 815 | 25 570 659 | 49.60% |
| Pre-parasitic J2 | B | 43 883 143 | 21 477 892 | 49.00% |
| Pre-parasitic J2 | C | 44 867 529 | 22 072 545 | 49.20% |
| Mc1-potato 8 dpi | A | 46 115 017 | 1 572 404 | 3.40% |
| Mc1-potato 8 dpi | B | 40 726 172 | 1 294 772 | 3.20% |
| Mc1-potato 8 dpi | C | 40 068 646 | 1 357 759 | 3.40% |
Information about the 11 M_ chitwoodi genes analyzed by qRT-PCR at different life stages_
| Gene ID (aa) | BLASTx nr | BLASTx % query cover | BLASTx e-value | Predicted domains | FPKM J2 | FPKM 8 dpi |
|---|---|---|---|---|---|---|
| Mc07118 (171) | Meloidogyne graminicola MSP18 mRNA, complete cds, Genbank: MK628546.1 | 84 | 7.00E-40 | N/A | 2 | 73.9 |
| Mc10102 (103) | Unnamed protein product [Meloidogyne enterolobii] Genbank: CAD2180454.1 | 79 | 4.00E-46 | Cytochrome b5, heme binding domain | 16.7 | 418.6 |
| Mc08299 (207) | Cathepsin B [Meloidogyne graminicola]Genbank KAF7630769.1 | 89 | 7.00E-45 | cysteine proteinase | 26.4 | 476 |
| Mc04921 (544) | hypothetical protein Mgra_00005223 [Meloidogyne graminicola] Genbank KAF7635403.1 | 34 | 6.00E-74 | Thioredoxin-like fold | 2.9 | 50.4 |
| Mc06869 (63) | N/A | N/A | 39.2 | 1704.9 | ||
| Mc03992 (306) | Hypothetical protein Mgra_00005223 [Meloidogyne graminicola] Genbank KAF76334147.1 | 49 | 3.20E-02 | N/A | 4.3 | 69.8 |
| Mc01565 (430) | Unnamed protein product [Meloidogyne enterolobii] Genbank: CAD2124200.1 | 18 | 2.00E-22 | N/A | 7.3 | 288.1 |
| Mc08895 (332) | Unnamed protein product [Meloidogyne enterolobii] Genbank: CAD2174631.1 | 32 | 5.00E-25 | N/A | 3.5 | 123.7 |
| Mc04677 (315) | Hypothetical protein Mgra_00005223 [Meloidogyne graminicola] Genbank KAF7635986.1 | 59 | 7.00E-54 | N/A | 2.3 | 51.8 |
| Mc03439 (79) | Hypothetical protein Mgra_00005223 [Meloidogyne graminicola] Genbank KAF7630992.1 | 97 | 1.00E-23 | N/A | 25.8 | 1319.5 |
| Mc10400 (130) | N/A | N/A | 7.2 | 154.8 |
Top 10 most differentially expressed genes in parasitic M_ chitwoodi juveniles_
| Log FC | Description | ||
| Up-regulated | 1 | 12.2878 | Putative cuticular collagen |
| 2 | 10.8863 | ---NA--- | |
| 3 | 10.6047 | Putative esophageal gland cell secretory protein 14 | |
| 4 | 10.5679 | ---NA--- | |
| 5 | 10.5294 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | |
| 6 | 9.2692 | ---NA--- | |
| 7 | 9.21531 | ---NA--- | |
| 8 | 9.21423 | Nematode cuticle collagen domain protein | |
| 9 | 9.1679 | Saposin-like type B, 1 domain and Saposin B domain and Saposin-like domain-containing protein | |
| 10 | 9.0144 | ---NA--- | |
| Down-regulated | 1 | -11.012 | C-type lectin domain-containing protein |
| 2 | -10.711 | Mucin-like protein-1 | |
| 3 | -10.681 | SCP domain-containing protein | |
| 4 | -10.443 | ---NA--- | |
| 5 | -10.308 | Beta-1,4-endoglucanase | |
| 6 | -10.234 | C-type lectin domain-containing protein | |
| 7 | -10.205 | Mucin-like protein-1 | |
| 8 | -9.9796 | ---NA--- | |
| 9 | -9.975 | ---NA--- | |
| 10 | -9.8201 | Hypothetical protein Mgra_00005110, partial |
Primers for qRT-PCR_
| Name | 5′-3′ |
|---|---|
| McITS2 forward | GGGGTCAAACCCTTTGGCACGTCTGG |
| McITS2 reverse | GCGGGTGATCTCGACTGAGTTCAGG |
| Mc00773 forward | AAGTTGCCGATAGCATTGCG |
| Mc00773 reverse | TCCGGAAGAGCATGACGAAT |
| Mc03439 forward | TGCAGTTGCTGAGAGTTGTC |
| Mc03439 reverse | TGCATGGTGGAATAACCAAAGT |
| Mc03992 forward | TGATCGTTCGACTTCTGGACA |
| Mc03992 reverse | GCACGAGTGCCTTGAACTTG |
| Mc04677 forward | CTGCACATCAGAAGAAAAATGCT |
| Mc04677 reverse | TCCACAGGGTCACAACTTCC |
| Mc04921 forward | TCGTGTTAGCGGTGATGGTT |
| Mc04921 reverse | CGTTTCGGTGCCAAGTTCAG |
| Mc08895 forward | TCTTTGCACGAAGTTGATCG |
| Mc08895 reverse | TTTAATCCATTTACAAAAAGCCCT |
| Mc10400 forward | CAAGGAGGTGGAAAAACGCC |
| Mc10400 reverse | GGGTTCTTGATTCCCAACCG |
| Mc01565 forward | CAGCTGAATCTTCTGCCCCA |
| Mc01565 reverse | AATAGCAACGGCTGGAGCTT |
| Mc06869 forward | CCACATCATCATCATCATTCAAGT |
| Mc06869 reverse | CCCATGGCCAAGTTGAACC |
| Mc08299 forward | GAATTACGCCGCTTTCTTGGT |
| Mc08299 reverse | TGCACATTCAGGCCACTCAT |
| Mc07118 forward | TGGTTGTTATGAACATTCTGGCG |
| Mc07118 reverse | CCAGGTTTGTTAGGTAAGGCAG |
| Mc10102 forward | TCCACATCATCCTGGAGGTC |
| Mc10102 reverse | ATGAAAAGCACCAGCCCCAT |