Fig. 1.
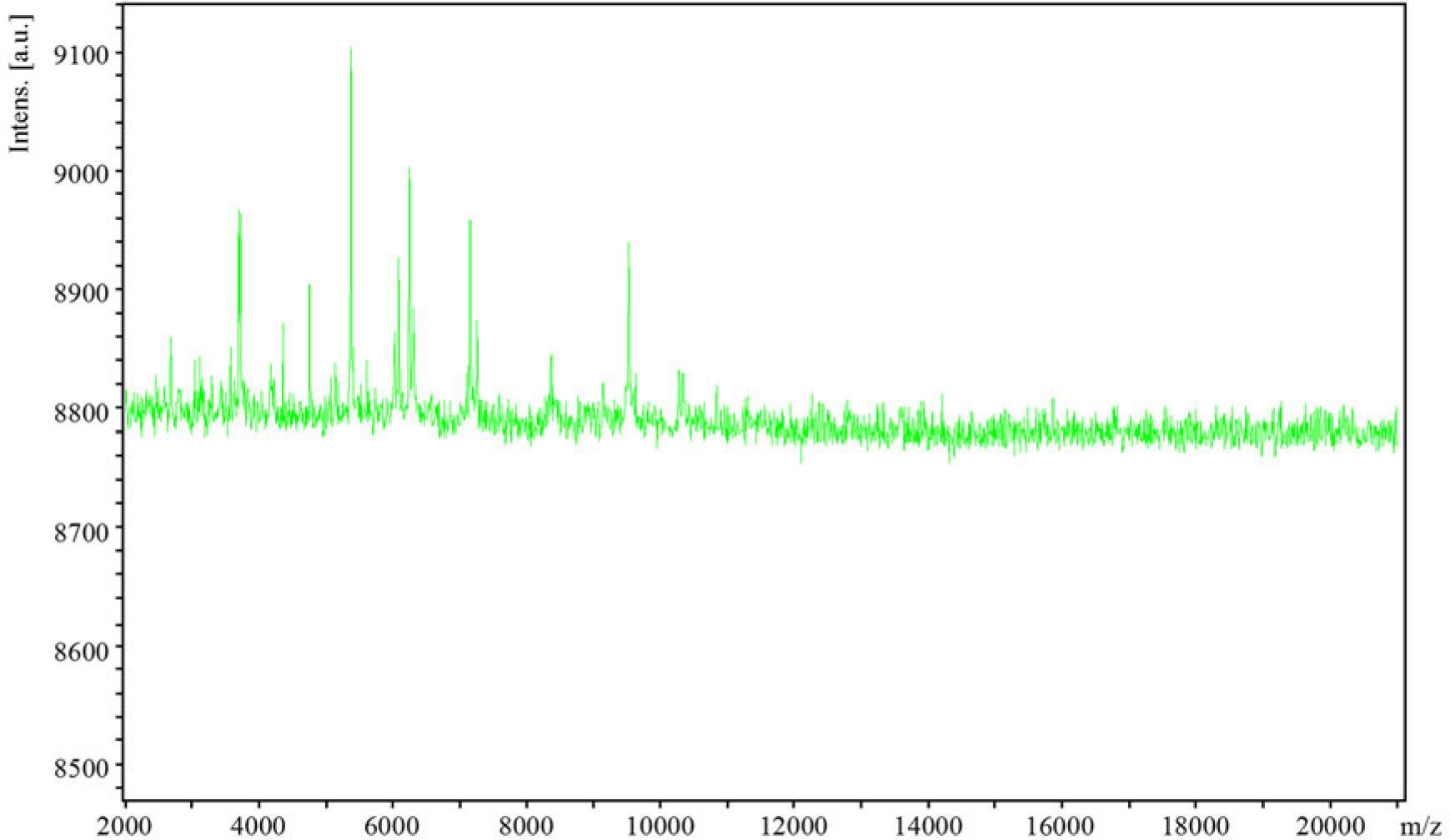
Fig. 2.
Fig. 3.
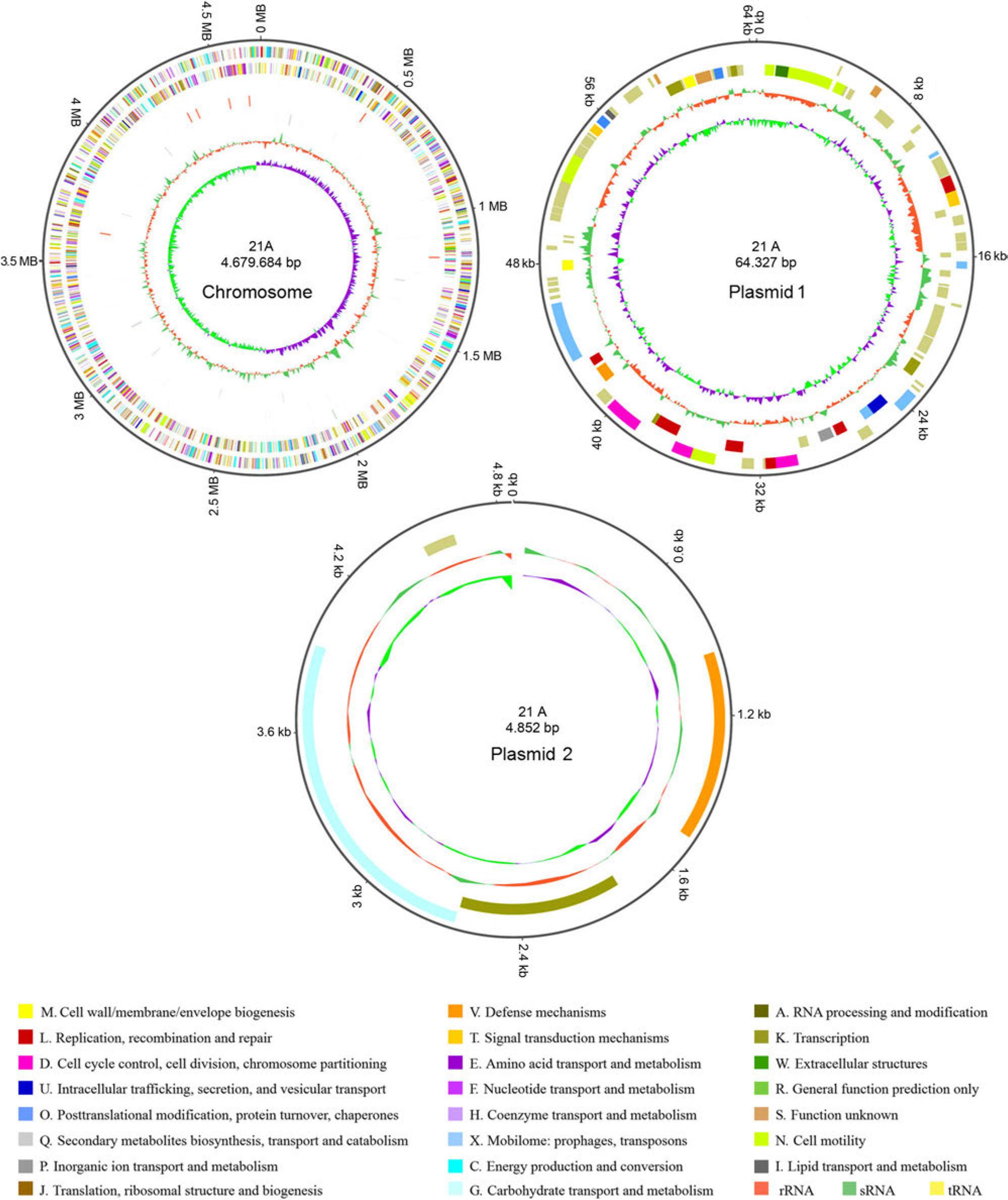
Fig. 4.
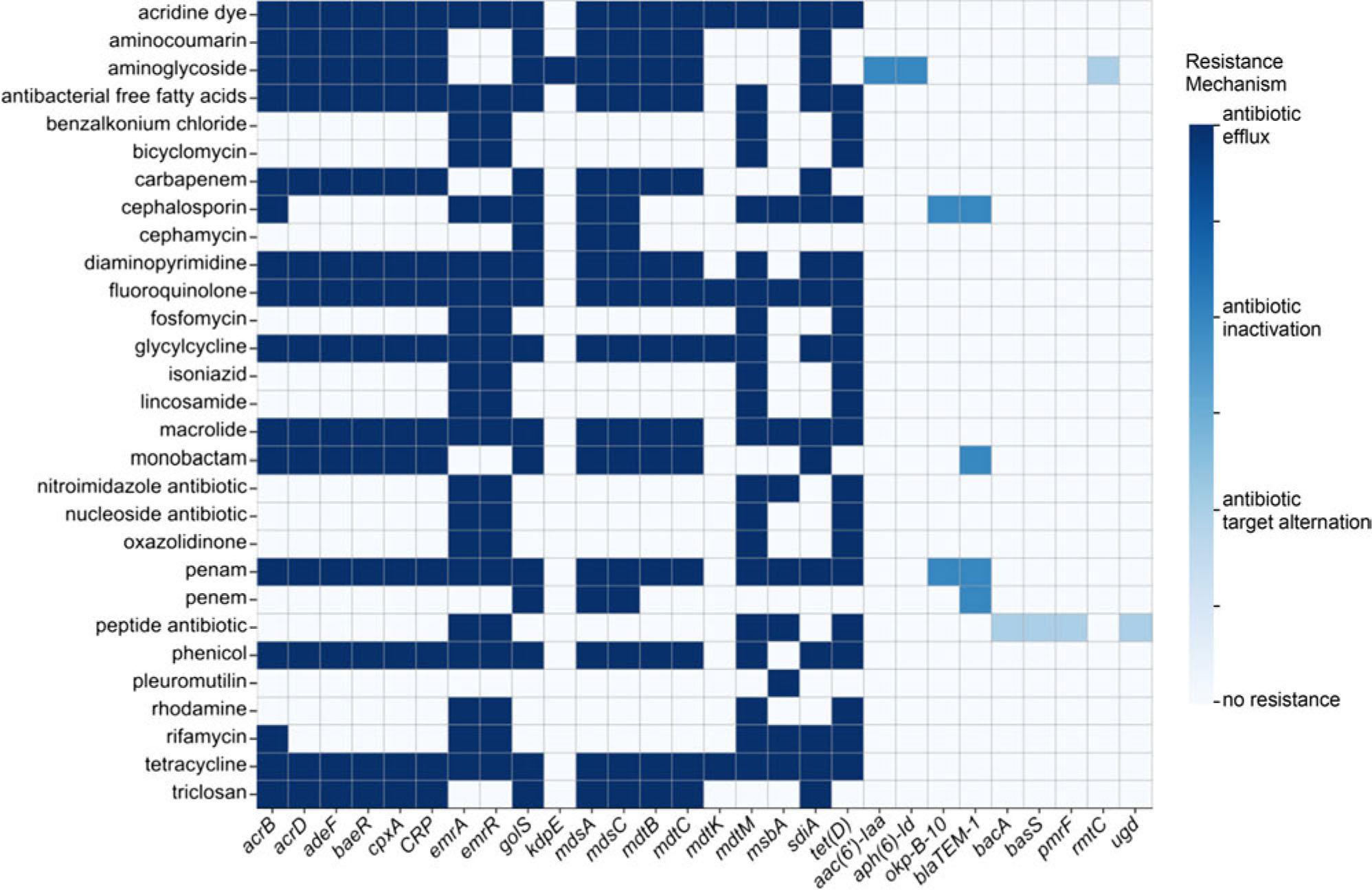
Fig. 5.
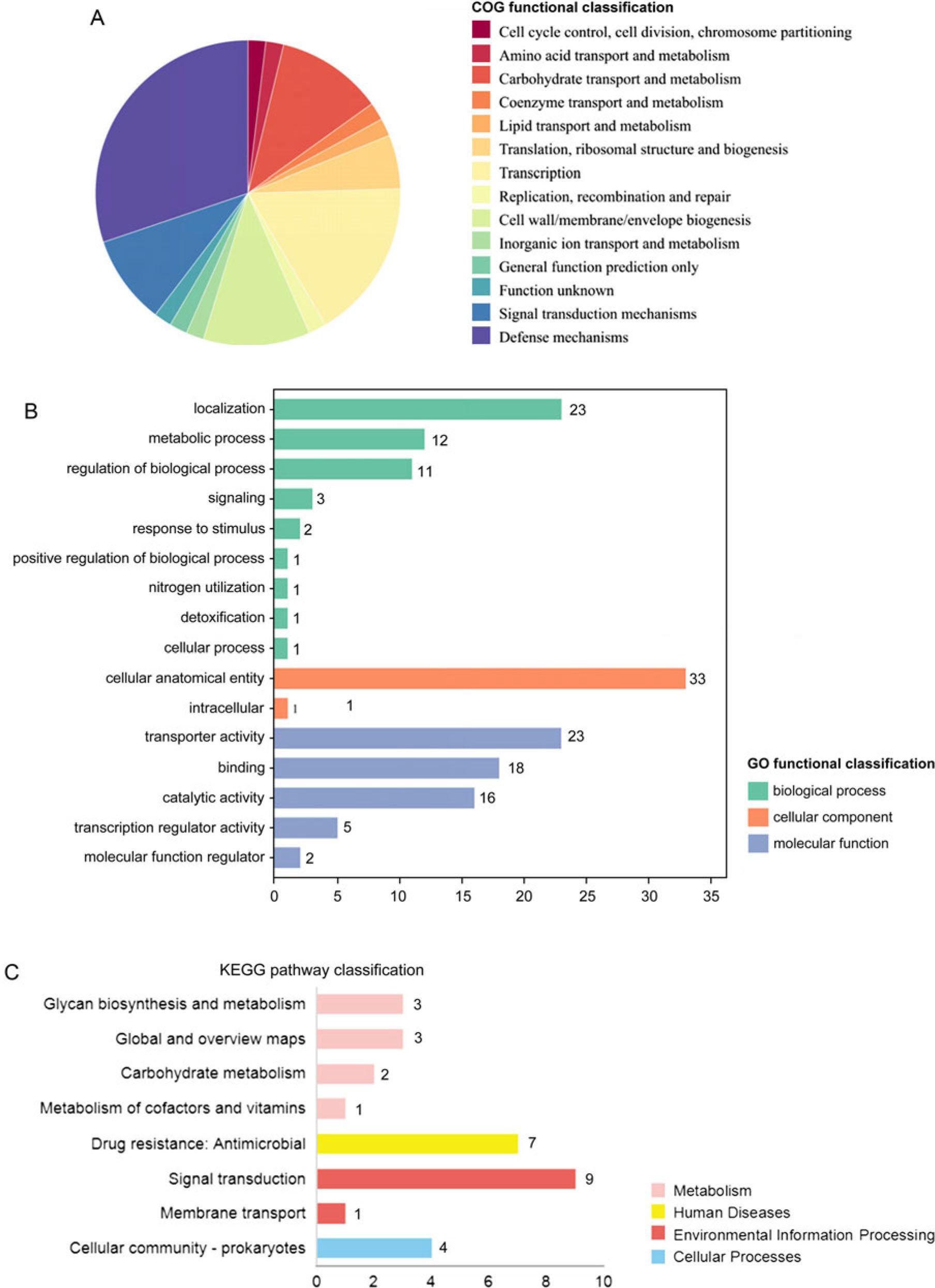
Fig. 6.
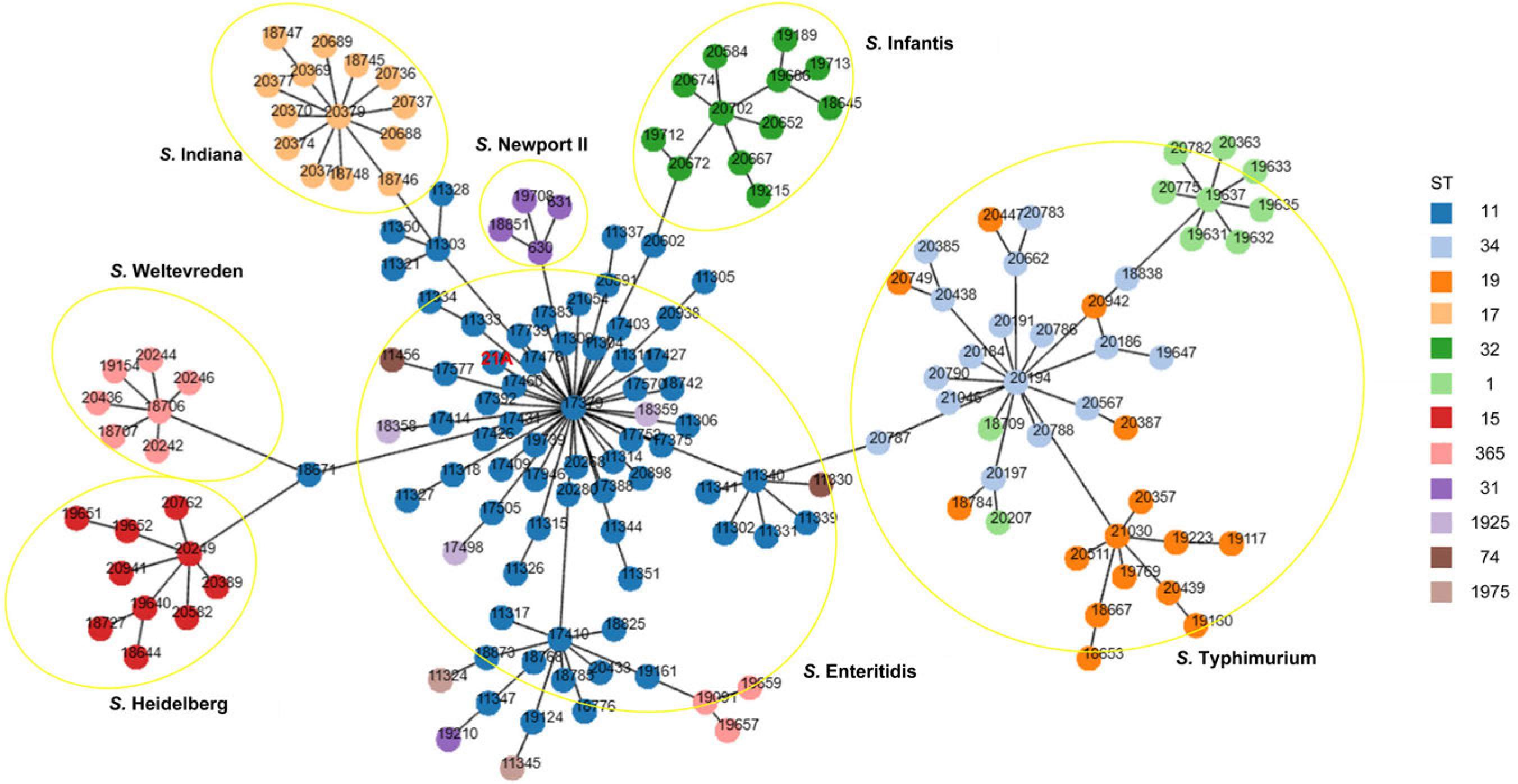
Fig. 7.
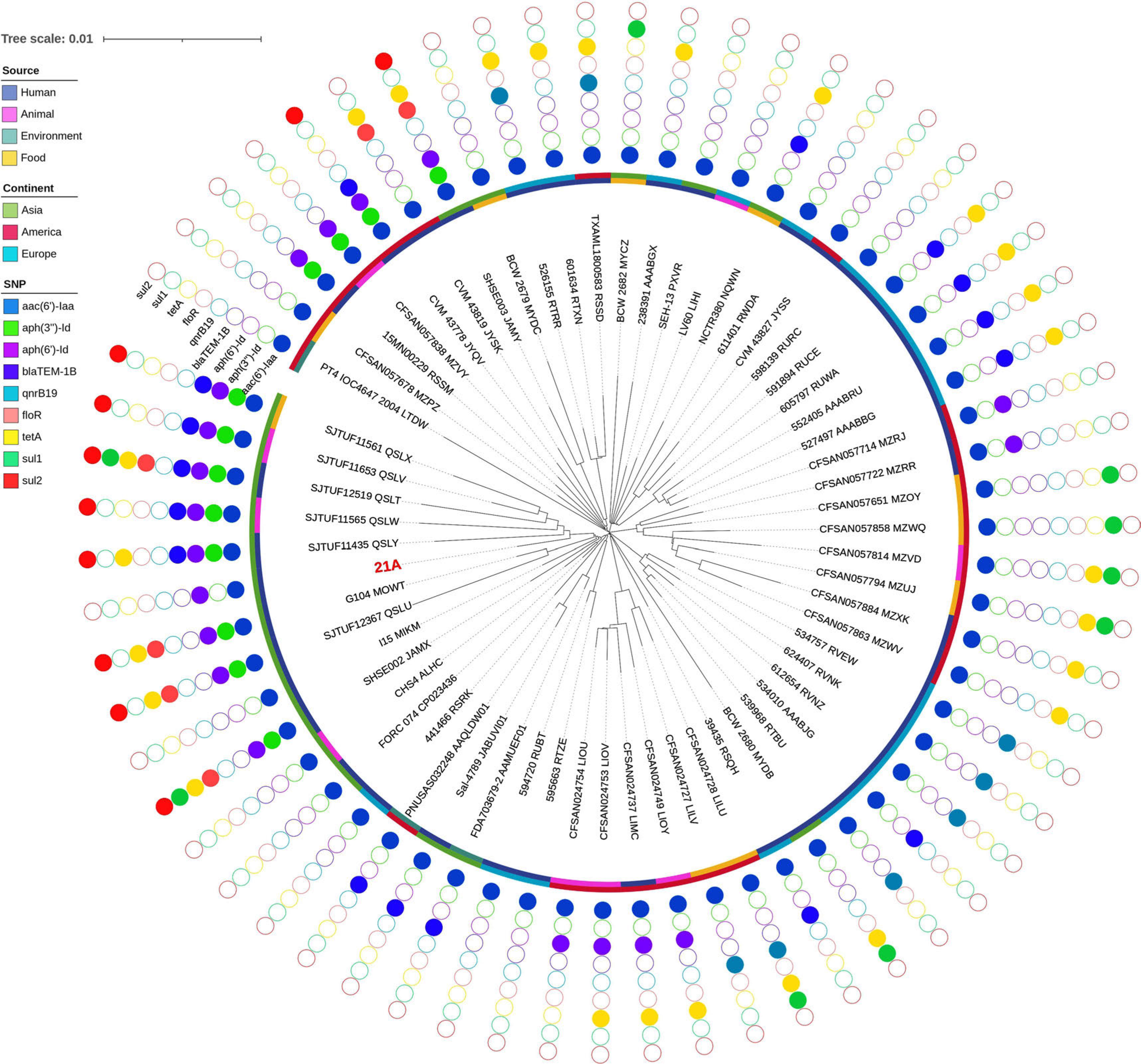
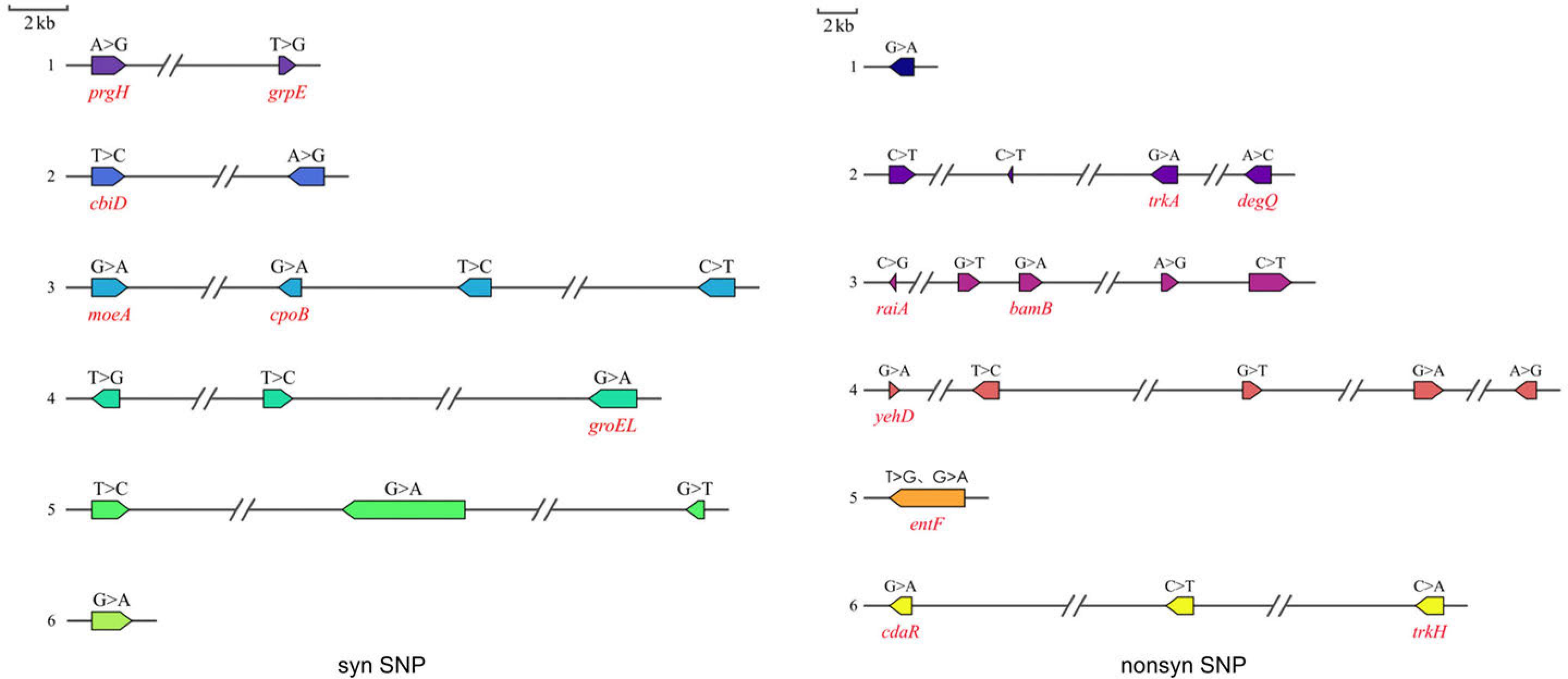
Fig. 8.
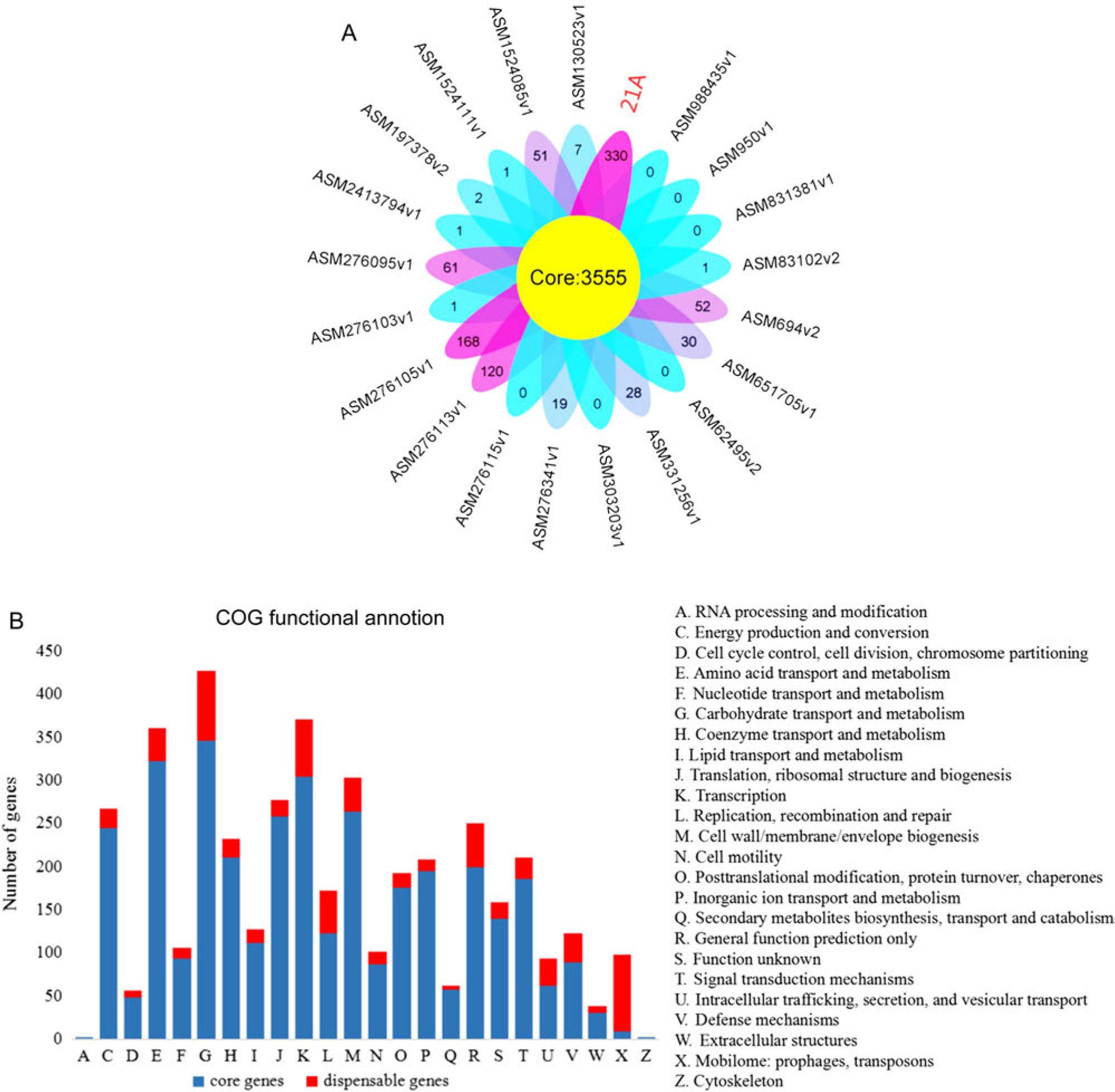
Fig. 9.
Matching scores of mass spectrometry identification results_
| Grade (quality) | Matching mode | Score | NCBI identifier |
|---|---|---|---|
| 1 (++) | Salmonella sp. (enterica st Hadar)Sa05 506 VAB | 2.030 | 149385 |
| 2 (++) | Salmonella sp. (enterica st Anatum)11 LAL | 2.023 | 58712 |
| 3 (+) | Salmonella sp. (choleraesuis)08 LAL | 1.998 | 591 |
| 4 (+) | Salmonella sp. (enterica st Dublin)Sa05 188 VAB | 1.972 | 98360 |
| 5 (+) | Salmonella sp. (enterica st Enterica)DSM 17058T HAM | 1.915 | 59201 |
| 6 (+) | Salmonella sp. (typhimurium)12 LAL | 1.894 | 602 |
| 7 (+) | Salmonella sp. (enteritidis)25089078 (PX)MLD | 1.885 | 592 |
| 8 (+) | Citrobacter koseri 9553-1 CHB | 1.783 | 545 |
| 9 (+) | Citrobacter koseri Mu15167-1 CHB | 1.753 | 545 |
| 10 (+) | Salmonella sp. (enterica st Stanley)15 LAL | 1.752 | 192953 |
VFDB database predicted the virulence factors of strain 21A_
| Type | Genes | |
|---|---|---|
| TTSS | SPI-1 | avrA, hilCD, iacP, iagB, invABCEFGHIJ, orgABC, prgHIJK, sicAP, sipABCD, sitABCD, sopABDEE2, spaOPQRS, sprB, sptP, steA |
| SPI-2 | pipBB2, sifAB, sopD2, ssaCDEGHIJKLMNOPQRTUV, sscAB, sseBCDEFGIJKL, sspH2, ssrAB, steC | |
| SPI-3 | mgtBC, misL | |
| SPI-4 | siiE | |
| T3SS | ABB77417, hopAN1, hrpH, mlr6326, RSp0731, spvABCDR | |
| Flagella and chemotaxis | Flagella | fleQ, fleR |
| Polar flagella | fleR/flrC, nueA, flrA, flmH | |
| Lateral flagella | lafK | |
| Peritrichous flagella | cheABDRWYZ, flgABCDEFGHIJKLMN, flhABCDE, fliABDEFGHIJKLMNOPQRSTYZ, fljB, flk, motAB, tar/cheM | |
| Fimbriae | Fimbrial subunit | bcfADEF, lpfE, pefA, pegA, safD, stbA, steADEF, stfAEFG, sthDE, stiA |
| Fimbrial chaperone | bcfBC, fimC, lpfB, safB, sefB, stbBE, stdC, steC, stiB | |
| Type IV pili | vfr, rpoN, pilRTW | |
| Other related genes | algU, bcfC, fimADFHIWYZ, lpfAD, pefB, pegC, safC, sefCR, stbCD, stdAB, steB, stfC, sthA, stiCH | |
| Lipopolysaccharide (LPS) | LOS | galU, gmhA, htrB, kdsA, lpxABCDHK, msbA, opsX, orfM, rfaDEF, wbaP, wecA |
| O-antigen | ddhACD, prt, wbcC | |
| Other related genes | acpXL, bplF, fabZ, gtrAB, hisH2, kdtB, orfH, pagP, waaGP | |
| Adhesins | bcfH, csgABCDEFG, lpfC, pagN, pefCD, stdD, stfD, sthBC, upaGH | |
| Capsule | cap8J, cpsF, galF, gmd, gnd, kpsF, manB, oppF, rmlAB, uppS, wbfBD, wcaHI, wcbN, wzb | |
| Efflux pump | adeFG, ccmB, fbpC, fepC, hitC, mtrE, phuU, sugC | |
| Others | hlyA, entABCDEFS, rck, sodB, sodCI, acrB, allABCDRS, bfmR, btpA, cesT, clpEP, fes, icl, mip, pvcC, ratB, ricA, sinH, cofT, virB, mig-5 | |
Basic genomic component information of strain 21A_
| Type | Number | Total length (bp) | GC content (%) | |
|---|---|---|---|---|
| Genome | 4663 | 4,118,712 | 53.34 | |
| Number | Total length (bp) | In genome (%) | ||
| ncRNA | tRNA | 21 | 1,647 | 0.0347 |
| 5S rRNA | 8 | 920 | 0.0193 | |
| 16S rRNA | 7 | 10,702 | 0.2253 | |
| 23S rRNA | 7 | 20,453 | 0.4306 | |
| sRNA | 68 | 7,926 | 0.1669 | |
| Tandem Repeat | TRF | 73 | 9,851 | 0.2074 |
| Minisatellite DNA | 50 | 3,611 | 0.076 | |
| Microsatellite DNA | 5 | 190 | 0.004 | |
| CRISPR | 2 | 1,155 | 0.0243 | |
| Prophage | 9 | 217,435 | 4.5787 | |
Resistant antimicrobial phenotype for strain 21A_
| Type of antibiotics | Drug | MIC | Resistance |
|---|---|---|---|
| β-lactam | Ampicillin | ≥ 32 μg/ml | R |
| Piperacillin | ≥ 128 μg/ml | R | |
| Ampicillin/sulbactam | ≥ 32 μg/ml | R | |
| Piperacillin/sulbactam | ≤ 4 μg/ml | S | |
| Cefoperazone | 25 mm | S | |
| Cefatriaxone | ≤ 1 μg/ml | S | |
| Ceftazidime | ≤ 1 μg/ml | S | |
| Cefepime | ≤ 1 μg/ml | S | |
| Aztreonam | ≤ 1 μg/ml | S | |
| Carbopenem | Imipenem | ≤ 1 μg/ml | S |
| Meropenem | ≤ 1 μg/ml | S | |
| Sulfonamide | Compound sulfamethoxazole | ≤ 20 μg/ml | S |
| Nitrofuran | Furadantin | ≥ 128 μg/ml | I |
| Quinolone | Levofloxacin | ≥ 8 μg/ml | R |
| Ciprofloxacin | ≥ 4 μg/ml | R |