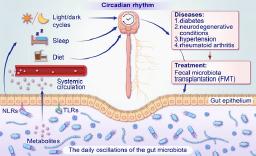
Fig. 1.
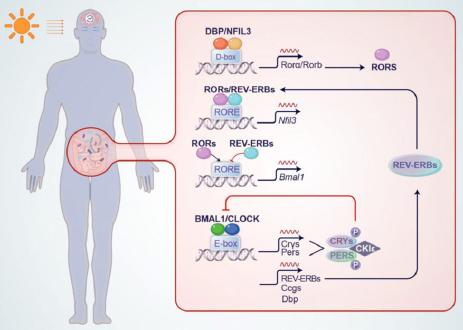
Evidences for bacterial circadian rhythms relevant to human health_
| Bacteria | Circadian components/time-dependent host response | References |
|---|---|---|
| Streptococcus pneumoniae | dependent on BMAL1 in Clara cells enhanced clearance at ZT12 | Gibbs et al. 2014 |
| Chlamydia trachomatis | enhanced clearance at ZT15 | Lundy et al. 2019 |
| Helicobacter pylori | enhanced lymphocyte migration to lymph nodes at ZT7 | Druzd et al. 2017 |
| Enterobacter aerogenes | swarming and motility rhythms | Paulose et al. 2016 |
| Pseudomonas putida | a putative kaiC homolog was found in the genome of Pseudomonas putida KT2440 by bioinformatics analyses | Soriano et al. 2010 |
| Escherichia coli | receptors for blue light | Gomelsky and Klug 2002 |
| Legionella pneumophila | kaiB and kaiC-encoding genes | Loza-Correa et al. 2010 |
| Salmonella Thyphimurium | enhanced clearance at ZT16 | Bellet et al. 2013 |
| Listeria monocytogenes | enhanced clearance at ZT8 dependent on BMAL1 in Ly6Chi monocytes | Nguyen et al. 2013 |