Fig. 1

Fig. 2
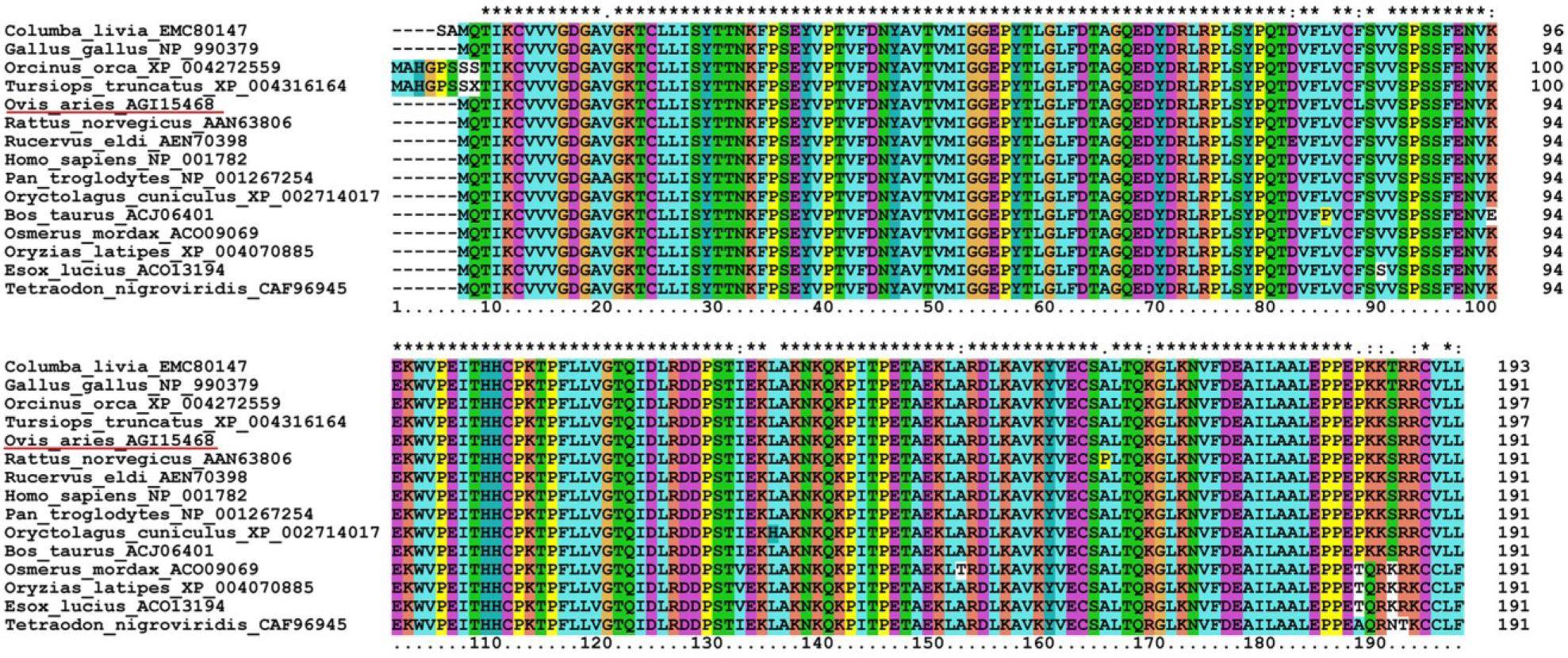
Fig. 3

Fig. 4
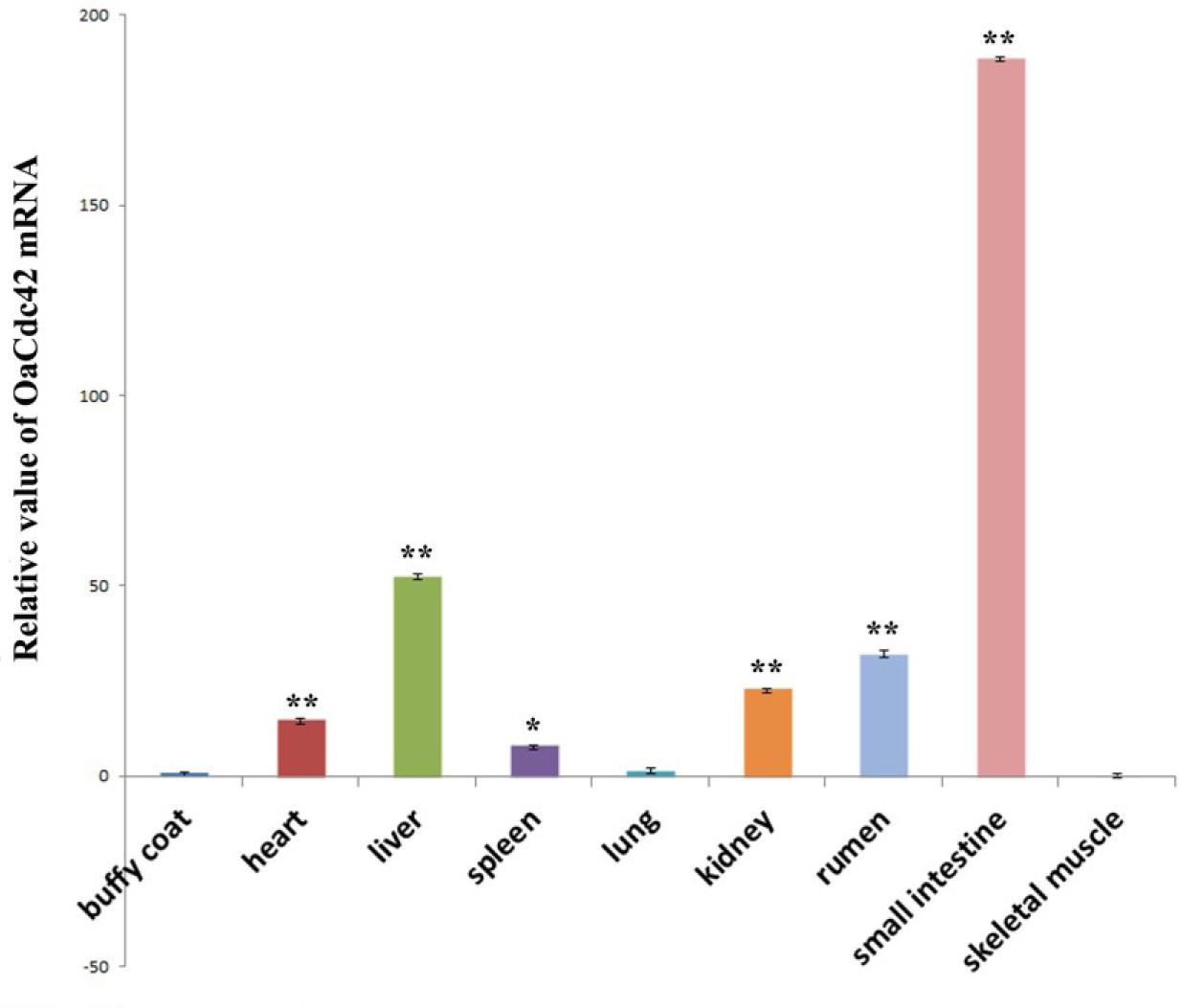
Fig. 5

Primers used for PCR amplification
| Primer | Nucleotide sequence (5’-3’) | Method |
|---|---|---|
| Cdc42 F1 | GTGTGAGACAAGGCCCGTAGGTATG | 3’- RACE; outer |
| Cdc42 F2 | TGGCCCCTTCCCCTCTCAATACTAG | 3’-RACE; inner |
| Cdc42 R1 | GCCATACCTACGGGCCTTGTCTCAC | 5’-RACE; outer |
| Cdc42 R2 | AGGTGCAGGGCATTTGTCATTATTG | 5’-RACE; inner |
| Cdc42 F’ | GTTGTTGTGGGTGATGGTGCTGTTG | Real-time PCR |
| Cdc42 R’ | CACTGAGAGGCAGACCAGAAACACG | Real-time PCR |
| β-actin F’ | CCCAAGGCCAACCGTGAGAAGATGA | Real-time PCR |
| β-actin R’ | CGAAGTCCAGGGCCACGTAGCAGAG | Real-time PCR |