Figure 1

Figure 2
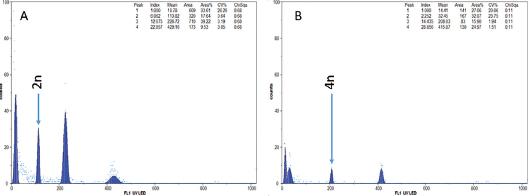
Figure 3

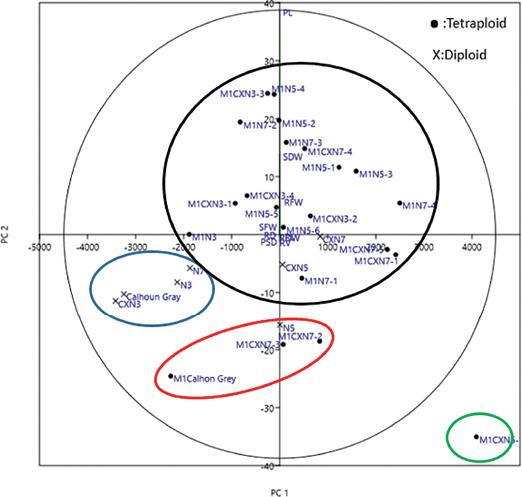
Figure 4

Figure 5
List, genotype codes and numbers of plant materials applied with colchicine_
| Genotype | Genotype code | Plants used in colchicine application (number) |
|---|---|---|
| Calhoun Gray (Citrullus lanatus) | C | 180 |
| W1482 (Citrullus lanatus var. citroides) | N3 | 210 |
| W1832 (Citrullus lanatus var. citroides) | N5 | 210 |
| W2001 (Citrullus lanatus var. citroides) | N7 | 180 |
| CxW1482 | CxN3 | 210 |
| CxW1832 | CxN5 | 180 |
| CxW2001 | CxN7 | 210 |
| Total | 1,380 |
Number of plants applied, the number of surviving plants and their ratio (%)_
| Genotype | Plants applied (number) | Surviving plants (number) | Surviving plants (%) |
|---|---|---|---|
| C | 180 | 120 | 66.67 |
| N3 | 210 | 119 | 56.67 |
| N5 | 210 | 110 | 52.38 |
| N7 | 180 | 158 | 87.78 |
| CXN3 | 210 | 116 | 55.24 |
| CXN5 | 180 | 119 | 66.11 |
| CXN7 | 210 | 116 | 55.24 |
| Total/mean | 1,380 | 858 | 62.17 |
RFW, RDW, RL, RV and RD of the plants grown in hydroponic culture for 21 days after transplanting_
| Genotype code | Ploidy level | RFW (g · plant-1) | RDW (g · plant-1) | RL (cm) | RV (cm3 · plant-1) | RD (mm) |
|---|---|---|---|---|---|---|
| M1Calhon Gray | Tetraploid | 9.81 mn | 1.15 pr | 1,319.16 m | 3.31 l–n | 0.40 g |
| M1N3 | Tetraploid | 10.50 l–n | 1.22 o–r | 1,702.86 m | 1.82 o | 0.37 i |
| M1N5-1 | Tetraploid | 22.13 fg | 2.26 h–k | 4,818.78 ef | 3.52 k–m | 0.44 f |
| M1N5-2 | Tetraploid | 22.94 d–f | 2.46 f–i | 3,570.95 g–k | 3.85 kl | 0.38 i |
| M1N5-3 | Tetraploid | 23.61 d–f | 2.53 e–h | 5,175.32 de | 5.92 gh | 0.53 c |
| M1N5-4 | Tetraploid | 26.78 c–e | 2.73 d–g | 3,470.66 h–l | 5.25 h–j | 0.44 f |
| M1N5-5 | Tetraploid | 20.26 f–h | 2.20 h–k | 3,514.55 h–k | 4.32 l | 0.40 g |
| M1N5-6 | Tetraploid | 23.48 d–f | 2.43 f–i | 3,660.05 g–j | 4.48 i–k | 0.58 a |
| M1N7-1 | Tetraploid | 13.97 k–m | 1.52 n–p | 4,046.80 f–i | 4.44 i–k | 0.37 i |
| M1N7-2 | Tetraploid | 14.51 j–l | 1.63 m–o | 2,764.20 kl | 3.95 kl | 0.55 b |
| M1N7-3 | Tetraploid | 20.65 f–h | 2.19 h–k | 3,722.38 g–j | 5.38 h–j | 0.44 f |
| M1N7-4 | Tetraploid | 21.48 fg | 2.33 g–j | 6,083.45 c | 2.81 mn | 0.37 i |
| M1CXN3-1 | Tetraploid | 15.73 i–k | 1.68 l–n | 2,658.33 l | 4.35 j–l | 0.46 e |
| M1CXN3-2 | Tetraploid | 28.37 c | 2.93 de | 4,223.48 f–h | 6.53 fg | 0.46 e |
| M1CXN3-3 | Tetraploid | 22.50 fg | 2.43 f–i | 3,333.78 i–l | 6.67 fg | 0.51 d |
| M1CXN3-4 | Tetraploid | 18.24 g–j | 1.93 j–n | 2,905.59 j–l | 3.79 k–m | 0.40 g |
| M1CXN5-1 | Tetraploid | 33.02 b | 3.41 bc | 7,678.59 b | 11.09 b | 0.44 f |
| M1CXN7-1 | Tetraploid | 27.09 cd | 2.86 d–f | 6,001.24 c | 7.40 d–f | 0.40 g |
| M1CXN7-2 | Tetraploid | 19.91 f–i | 2.05 i–m | 4,416.07 e–g | 4.50 i–k | 0.35 j |
| M1CXN7-3 | Tetraploid | 20.60 f–h | 2.23 h–k | 3,658.58 g–j | 7.28 ef | 0.51 d |
| M1CXN7-4 | Tetraploid | 28.94 c | 3.04 c–e | 4,104.75 f–i | 8.21 de | 0.51 d |
| M1CXN7-5 | Tetraploid | 22.82 d–f | 2.45 f–i | 5,825.66 cd | 8.29 d | 0.44 f |
| Calhoun Gray | Diploid | 3.16 o | 0.48 s | 344.85 n | 0.78 p | 0.44 f |
| CXN3 | Diploid | 1.70 o | 0.22 s | 173.10 n | 0.27 p | 0.46 e |
| CXN5 | Diploid | 19.29 f–i | 2.09 h–l | 3,645.02 g–j | 4.22 l | 0.40 g |
| CXN7 | Diploid | 16.70 h–k | 1.83 k–n | 4,428.47 e–g | 5.51 hi | 0.40 g |
| N3 | Diploid | 8.37 n | 0.89 r | 1,454.43 m | 1.82 o | 0.40 g |
| N5 | Diploid | 13.90 k–n | 1.57 n–p | 3,591.94 g–k | 4.31 l | 0.40 g |
| N7 | Diploid | 10.77 n | 1.16 pr | 1,719.21 m | 2.44 no | 0.44 f |
| C. tide | Citrullus lanatus | 1.19 o | 0.24 s | 148.66 n | 0.32 p | 0.37 i |
| RS841 | C. maxima × C. moschata | 38.17 a | 4.00 a | 11,098.84 a | 13.04 a | 0.39 h |
| Argentario | Lagenaria siceraria | 34.00 b | 3.50 b | 11,000.44 a | 9.70 c | 0.34 k |
Mean stomatal diameter, stomatal length, stomatal density and chloroplast number in presumed polyploid and diploid plants_
| Stomatal diameter (μm) | Stomatal length (μm) | Stomatal density (number · mm-2) | Chloroplast number (number) | |||||
|---|---|---|---|---|---|---|---|---|
| Ploidy level | Diploid | Tetraploid | Diploid | Tetraploid | Diploid | Tetraploid | Diploid | Tetraploid |
| Maximum | 16.09 | 20.56 | 23.30 | 31.42 | 627.30 | 341.75 | 10.18 | 16.41 |
| Minimum | 18.90 | 23.87 | 26.04 | 34.01 | 927.77 | 470.34 | 12.03 | 19.37 |
| Mean | 17.50 | 22.22 | 24.67 | 32.72 | 777.54 | 406.05 | 11.11 | 17.89 |
The number and percentage of tetraploid plants obtained as a result of microscope and flow cytometry analyses_
| Genotype | Number of tetraploid plants | Flow cytometry analysis percentage of tetraploid plants (%) | |
|---|---|---|---|
| Stomatal examinations (number) | Flow cytometry analysis | ||
| C | 8 | 1 | 12.50 |
| N3 | 15 | 1 | 6.67 |
| N5 | 11 | 6 | 54.55 |
| N7 | 12 | 4 | 33.33 |
| CXN3 | 14 | 4 | 28.57 |
| CXN5 | 11 | 1 | 9.09 |
| CXN7 | 13 | 5 | 38.46 |
| Total/mean | 84 | 22 | 26.19 |
Pearson’s correlation coefficients (r values) between DNA content and stomatal measurement in 22 tetraploid and 62 diploid watermelon genotypes_
| Stomatal diameter | Stomatal length | Stomatal density | Chloroplast number | |
|---|---|---|---|---|
| DNA content | 0.96** | 0.90** | -0.79** | 0.88** |
List of genotypes used in hydroponic culture_
| Genotype code | Ploidy level | Genotype code | Ploidy level |
|---|---|---|---|
| M1Calhon Gray | Tetraploid | M1CXN5-1 | Tetraploid |
| M1N3 | Tetraploid | M1CXN7-1 | Tetraploid |
| M1N5-1 | Tetraploid | M1CXN7-2 | Tetraploid |
| M1N5-2 | Tetraploid | M1CXN7-3 | Tetraploid |
| M1N5-3 | Tetraploid | M1CXN7-4 | Tetraploid |
| M1N5-4 | Tetraploid | M1CXN7-5 | Tetraploid |
| M1N5-5 | Tetraploid | Calhoun Gray | Diploid |
| M1N5-6 | Tetraploid | CXN3 | Diploid |
| M1N7-1 | Tetraploid | CXN5 | Diploid |
| M1N7-2 | Tetraploid | CXN7 | Diploid |
| M1N7-3 | Tetraploid | N3 | Diploid |
| M1N7-4 | Tetraploid | N5 | Diploid |
| M1CXN3-1 | Tetraploid | N7 | Diploid |
| M1CXN3-2 | Tetraploid | Crimson Tide | Diploid |
| M1CXN3-3 | Tetraploid | RS841 | Diploid |
| M1CXN3-4 | Tetraploid | Argentario | Diploid |
PSD, plant height, SFW and SDW of the plants grown in hydroponic culture_
| Genotype code | Ploidy level | PSD (mm) | PL (cm) | SFW (g · plant-1) | SDW (g · plant-1) |
|---|---|---|---|---|---|
| M1Calhon Gray | Tetraploid | 5.82 b–d | 30.83 mn | 12.58 no | 1.42 op |
| M1N3 | Tetraploid | 5.61 b–d | 35.53 k–m | 21.37 k–m | 2.30 l–n |
| M1N5-1 | Tetraploid | 5.53 b–d | 67.40 c–f | 49.14 de | 4.96 d–f |
| M1N5-2 | Tetraploid | 5.57 b–d | 67.67 c–f | 37.50 f–i | 3.92 g–j |
| M1N5-3 | Tetraploid | 5.17 b–d | 73.67 cd | 39.17 f–h | 4.08 g–i |
| M1N5-4 | Tetraploid | 5.23 b–d | 70.33 cd | 39.17 f–h | 3.97 g–j |
| M1N5-5 | Tetraploid | 5.26 b–d | 54.00 e–j | 30.33 h–j | 3.21 i–k |
| M1N5-6 | Tetraploid | 4.94 b–d | 51.12 f–k | 30.83 h–j | 3.16 jl |
| M1N7-1 | Tetraploid | 5.32 b–d | 49.70 g–l | 23.33 j–m | 2.45 k–n |
| M1N7-2 | Tetraploid | 4.88 cd | 65.05 c–g | 25.00 j–m | 2.68 k–m |
| M1N7-3 | Tetraploid | 5.19 b–d | 64.25 c–h | 40.83 e–g | 4.21 f–h |
| M1N7-4 | Tetraploid | 5.38 b–d | 75.65 c | 43.33 e–g | 4.51 e–h |
| M1CXN3-1 | Tetraploid | 6.05 b–d | 46.67 i–m | 30.83 h–j | 3.19 jk |
| M1CXN3-2 | Tetraploid | 5.90 b–d | 48.18 h–l | 58.33 c | 5.93 c |
| M1CXN3-3 | Tetraploid | 5.77 b–d | 67.67 c–f | 45.00 e–g | 4.68 e–h |
| M1CXN3-4 | Tetraploid | 6.13 b–d | 52.02 f–k | 25.83 j–l | 2.69 k–m |
| M1CXN5-1 | Tetraploid | 6.48 b–d | 45.47 j–m | 46.33 ef | 4.75 e–g |
| M1CXN7-1 | Tetraploid | 6.12 b–d | 64.00 c–h | 44.67 e–g | 4.62 e–h |
| M1CXN7-2 | Tetraploid | 5.93 b–d | 34.55 l–n | 40.17 fg | 4.07 g–i |
| M1CXN7-3 | Tetraploid | 5.82 b–d | 27.67 no | 36.35 g–i | 3.81 h–j |
| M1CXN7-4 | Tetraploid | 6.05 b–d | 60.62 c–j | 55.00 cd | 5.65 cd |
| M1CXN7-5 | Tetraploid | 6.53 b–d | 62.17 c–i | 49.33 de | 5.10 d–f |
| Calhoun Gray | Diploid | 6.07 b–d | 17.83 o–r | 5.08 op | 0.67 pr |
| CXN3 | Diploid | 3.86 d | 15.55 o–r | 3.60 p | 0.41 r |
| CXN5 | Diploid | 5.48 b–d | 44.67 j–m | 30.83 h–j | 3.25 i–k |
| CXN7 | Diploid | 5.07 b–d | 58.52 d–j | 29.67 k | 3.13 jl |
| N3 | Diploid | 4.76 d | 26.22 n–p | 16.52 mn | 1.70 no |
| N5 | Diploid | 4.96 b-d | 37.23 k–n | 20.82 mn | 2.26 mn |
| N7 | Diploid | 4.11 d | 30.83 mn | 17.33 l–n | 1.81 no |
| C. tide | Citrullus lanatus | 2.79 d | 11.00 p–r | 2.72 p | 0.39 r |
| RS841 | C.maxima × C. moschata | 9.73 a | 95.83 b | 84.00 a | 8.58 a |
| Argentario | Lagenaria siceraria | 9.62 ab | 115.83 a | 73.33 b | 7.43 b |
Pearson’s correlation coefficients (r values) between plant growth parameters and direct (DNA content) and indirect (stomatal measurement) ploidy determination methods in 22 tetraploid and seven diploid watermelon genotypes_
| PSD | PL | SFW | SDW | RFW | RDW | RL | RV | RD | |
|---|---|---|---|---|---|---|---|---|---|
| DNA content | 0.33* | 0.48* | 0.60** | 0.60** | 0.60** | 0.61** | 0.45* | 0.47* | -0.02ns |
| Stomatal diameter | –0.15ns | –0.49* | –0.56** | –0.56** | -0.58** | -0.58** | -0.53** | -0.44* | 0.26ns |
| Stomatal length | 0.26ns | 0.50** | 0.49* | 0.49* | 0.52* | 0.53* | 0.38* | 0.32* | -0.15ns |
| Stomatal density | 0.30ns | 0.50** | 0.55** | 0.55** | 0.59** | 0.59** | 0.43* | 0.40* | -0.12ns |
| Chloroplast number | –0.19ns | 0.19ns | –0.10 ns | –0.10 ns | 0.02ns | 0.01ns | -0.05ns | -0.24ns | -0.26ns |
Thousand seed weights, germination rate, cotyledon width, cotyledon length, stem diameter and % change in genome size and 1,000 seed weights_
| Ploidy level | % Change | ||
|---|---|---|---|
| Tetraploid | Diploid | ||
| 1,000 seed weight (g) | 162.67 | 144.12 | 11.40 |
| Germination rate (%) | 65.8 | 94.8 | -30.59 |
| Cotyledon width (mm) | 27.92 | 25.89 | 7.76 |
| Cotyledon length (mm) | 39.99 | 35.53 | 12.58 |
| Hypocotyl diameter (mm) | 3.14 | 2.57 | 20.62 |