Figure 1
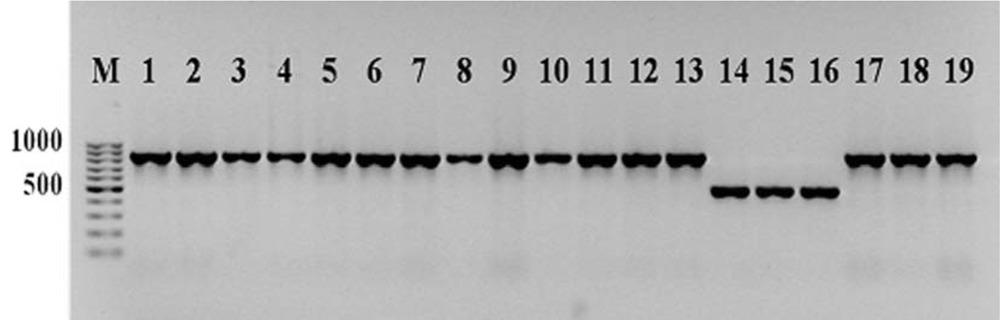
Figure 2
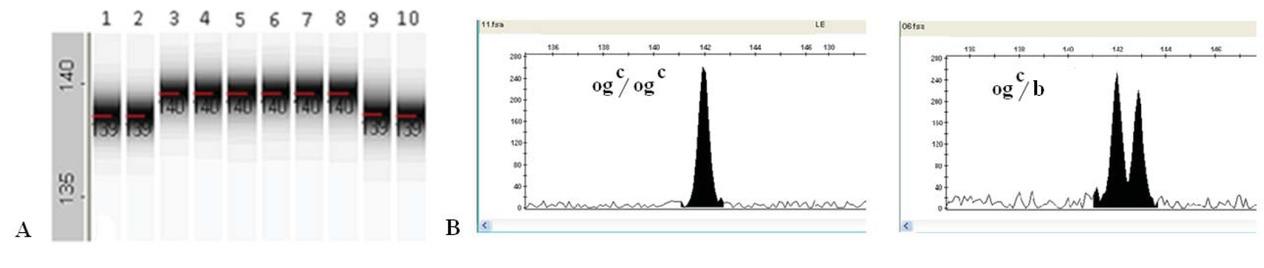
Figure 3
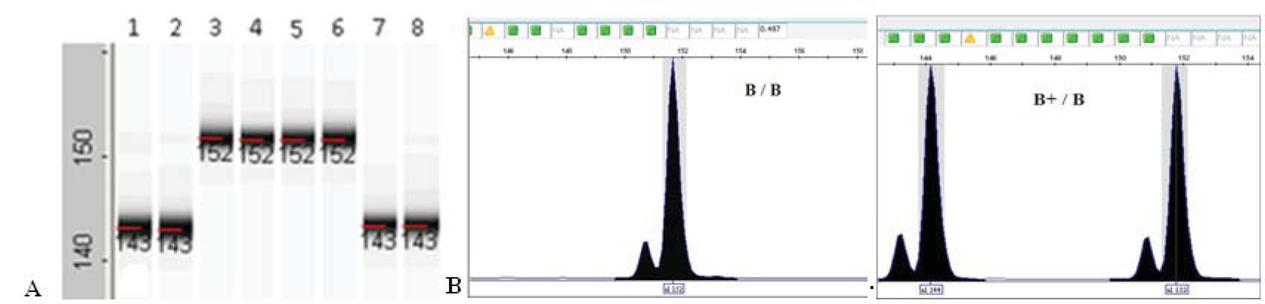
Figure 4
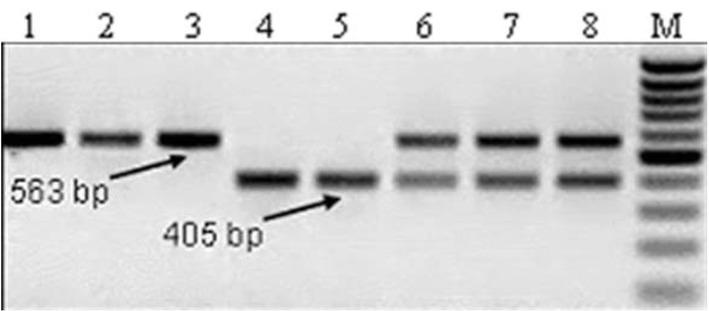
Figure 5
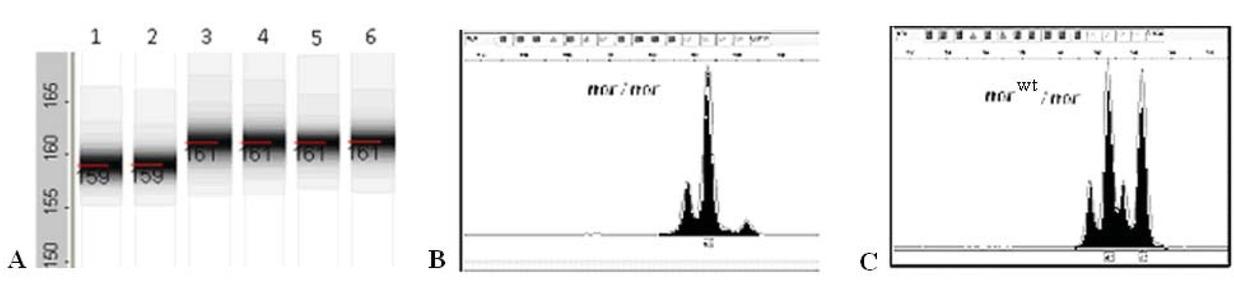
Figure 6

Figure 7
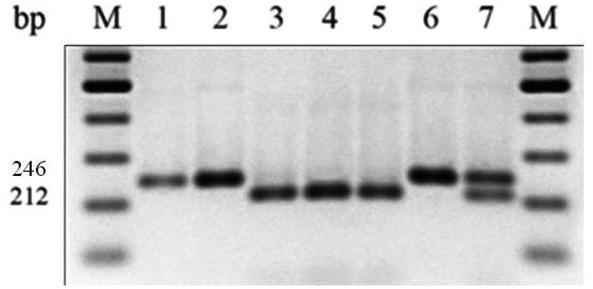
Figure 8
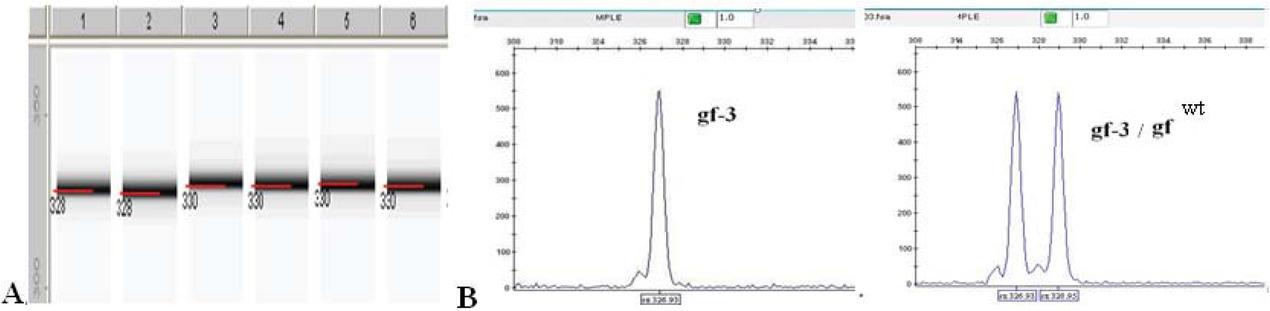
Figure 9
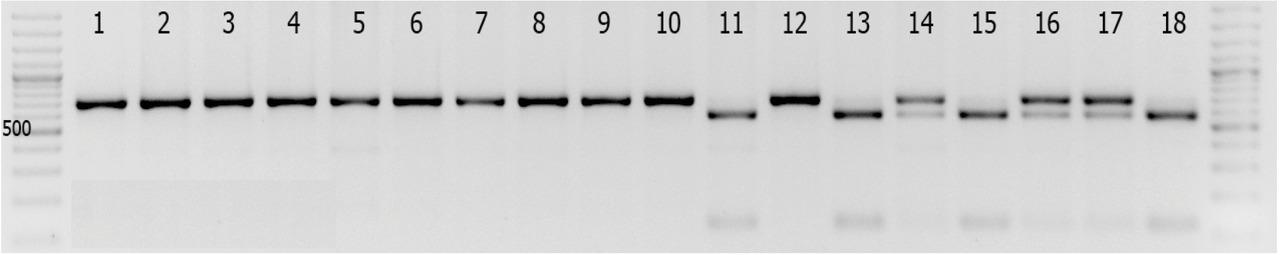
Figure 10

Figure 11
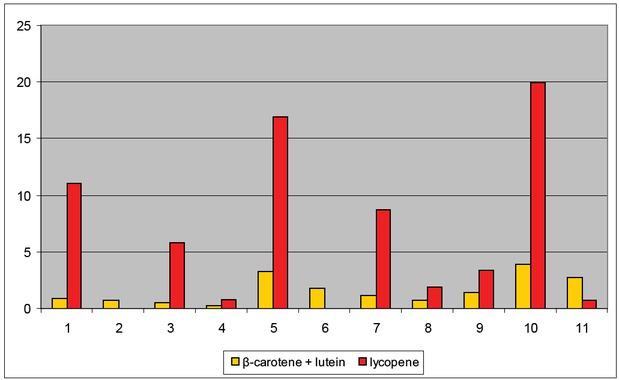
Figure 12
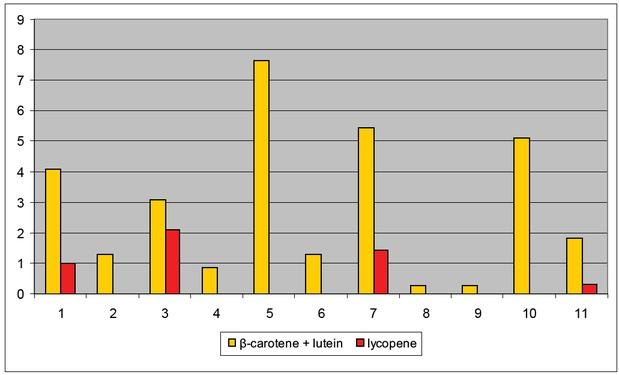
Carotenoid content in tomato samples with tangerine and non-ripening alleles (mg/100 g wet basis), 2016
| Allele combinations | neurosporin | ζ - carotene |
|---|---|---|
| t/t//norwt/norwt | 11.6 | 0 |
| t/t//norA/norA | 0 | 7.2 |
| t/t//nor/nor | 1.94 | 0.94 |
The content of carotenoids in tomato samples with different combinations of quality alleles (mg/100 g wet basis), 2016
| Allele combinations | β- carotene | Lycopene | Lutein | Chlorophyll | |
|---|---|---|---|---|---|
| a | b | ||||
| ogc/ogc//rinwt/rinwt | 0.85 | 11.0 | 0 | 0 | 0 |
| ogc/ogc//rin/rin | 0.44 | 0 | 0.26 | 0 | 0 |
| ogc/ogc//gf-3/gf-3//rinwt/rinwt | 0.55 | 5.75 | 0 | 0 | 0 |
| ogc/ogc//gf-3/gf-3//rin/rin | 0,25 | 0.8 | 0 | 0 | 0 |
| ogc/ogc//hp2dg/hp2 dg//rinwt/rinwt | 1.05 | 16.9 | 2.2 | 0 | 0 |
| ogc/ogc//hp2dg/hp2 dg//rin/rin | 0.7 | 0 | 1.1 | 0.60 | 0.62 |
| ogc/ogc//norwt/norwt | 1.15 | 8.7 | 0 | 0 | 0 |
| ogc/ogc//nor/nor | 0.7 | 1.9 | 0 | 0 | 0 |
| ogc/ogc//gf-3/gf-3//nor/nor | 0.83 | 3.4 | 0.57 | 0.28 | 0.28 |
| ogc/ogc//hp2dg/hp2dg//norwt/norwt | 1.7 | 19.9 | 2.2 | 0 | 0 |
| ogc/ogc//hp2dg/hp2dg//nor/nor | 1.4 | 0.7 | 1.4 | 0 | 0 |
| B/B//rinwt/rinwt | 2.9 | 1.0 | 1.2 | 0 | 0 |
| B/B//rin/rin | 0.69 | 0 | 0.6 | 0 | 0 |
| B/B//gf-3/gf-3//rinwt/rinwt | 3.1 | 2.1 | 0 | 0 | 0 |
| B/B//gf-3/gf-3//rin/rin | 0.6 | 0 | 0.25 | 0.87 | 0.93 |
| B/B//hp2dg/hp2 dg//rinwt/rinwt | 5.5 | 0 | 2.15 | 0 | 0 |
| B/B//hp2dg/hp2 dg//rin/rin | 0.6 | 0 | 0.7 | 0.68 | 0.63 |
| B/B//norwt/norwt | 4.7 | 1.43 | 0.75 | 0 | 0 |
| B/B//gf-3/gf-3//norwt/norwt | 1.8 | 5.5 | 1.0 | 3.0 | 2.1 |
| B/B//gf-3/gf-3//nor/nor | 0.28 | 0 | 0 | 2.3 | 2.6 |
| B/B//hp2dg/hp2 dg//norwt/norwt | 5.1 | 0 | 1.0 | 0 | 0 |
| B/B//hp2dg/hp2 dg//nor/nor | 1.63 | 0.3 | 1.2 | 1.01 | 0.80 |