Figure 1
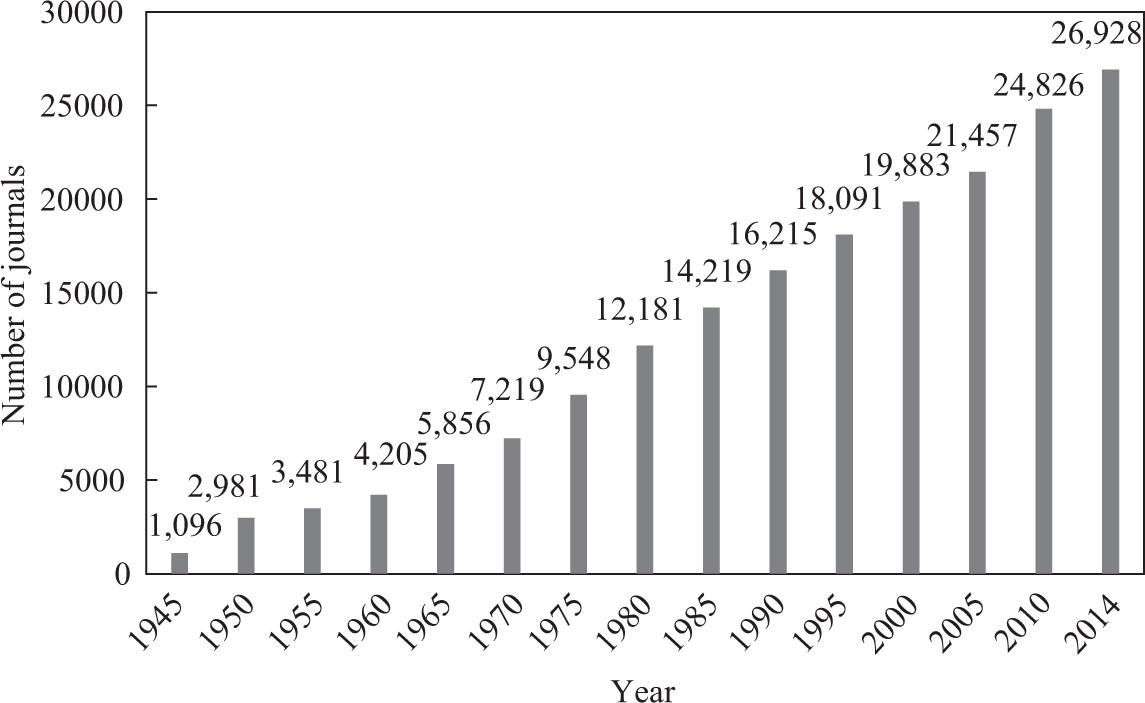
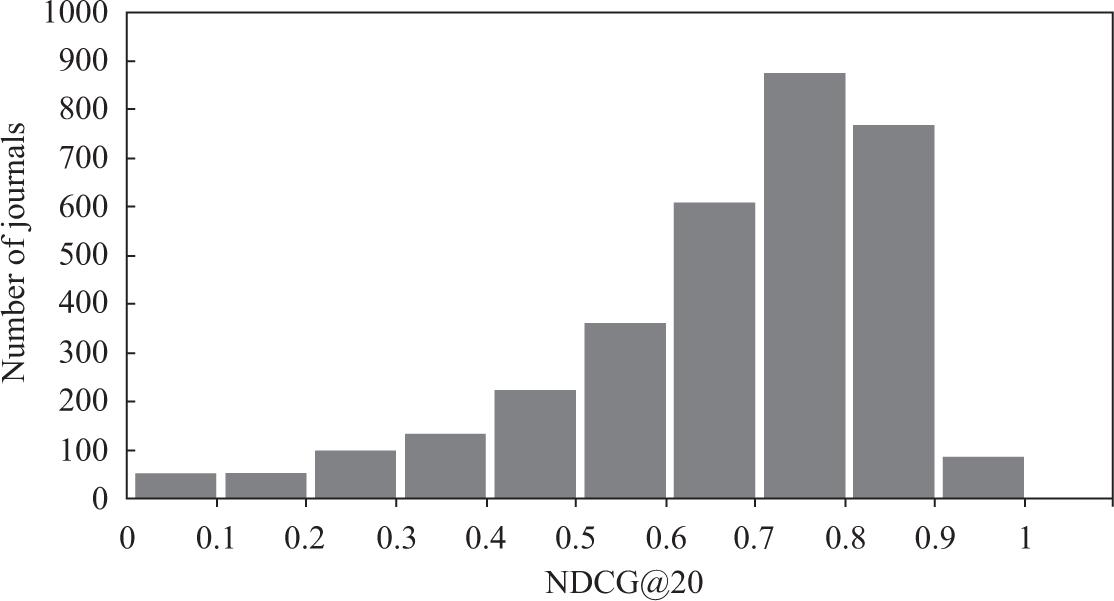
Figure 2
Top 3 MeSH terms in the articles of the journals identified by the three methods_
| Journal | MeSH #1 | MeSH #2 | MeSH #3 |
|---|---|---|---|
| Genetics | Mutation | Models, genetic | Genome |
| PNAS | DNA | Gene expression | Molecular sequence data |
| Cell | RNA | DNA | Molecular sequence data |
| Nature | Research | Research personnel | DNA |
| MCB | Gene expression | Cell line | Gene expression regulation |
| Science | DNA | Science | RNA |
| TAG | Chromosome mapping | Quantitative trait loci | Genes |
| Evolution | Evolution, molecular | Biological evolution | Selection, genetic |
| EMBO Journal | DNA | RNA | Gene expression |
| Genes & Development | Gene expression | Gene expression regulation | Cell line |
| NAR | DNA | RNA | Internet |
| JBC | Proteins | Gene expression regulation | Protein binding |
| MBE | Evolution, molecular | Phylogeny | Genome |
| Journal of Bacteriology | Bacteria | Gene expression | Gene expression regulation |
| MGG | Gene expression | Gene expression regulation | DNA |
| Heredity | Genetic variation | Genetics | Genetics, population |
| Development | Gene expression | Gene expression regulation | Gene expression regulation, developmental |
| Genetical Research | Models, genetic | Genotype | Chromosomes mapping |
| Genome | Phylogeny | Genes | Chromosomes |
| JMB | Models, molecular | Protein binding | Protein confirmation |
| JCB | Cell line | Protein transport | Cells |
| Current Biology: CB | Drosophila | Biological evolution | Gene expression |
| MBoC | Protein transport | RNA | Protein binding |
| Developmental Biology | Gene expression | Gene expression regulation | Gene expression regulation, developmental |
| Nature Genetics | Genome | Mutation | Polymorphism, single nucleotide |
| CCM | Intensive care | Intensive care units | Critical illness |
| PLOS Genetics | Gene expression | Gene expression regulation | DNA |
| AJHG | Mutation | Genetic predisposition to disease | Pedigree |
| PLOS One | Gene expression | Gene expression regulation | Cell line |
| Genome Research | Genome | Gene expression | DNA |
| Eukaryotic Cell | Fungal proteins | Gene expression | Gene expression regulation |
| Molecular Ecology | Genetic variation | Genetics, population | Genetics |
Top 10 journals related to JAMIA identified by the content-similarity-based approach_
| Journal | Broad subject term(s) |
|---|---|
| AMIA Annual Symposium Proceedings AMIA: The American Medical Informatics Association. AMIA Annual Symposium Proceedings is an ejournal published by AMIA annually. | Medical Informatics |
| BMC Medical Informatics and Decision Making | Medical Informatics |
| Journal of Biomedical Informatics | Medical Informatics |
| Studies in Health Technology and Informatics | Health Services Research |
| Medical Informatics | |
| Technology | |
| International Journal of Medical Informatics | Medical Informatics |
| BMC Bioinformatics | Computational Biology |
| Journal of Medical Internet Research | Medical Informatics |
| Health Technology Assessment | Health Services Research |
| Technology | |
| Journal of General Internal Medicine | Internal Medicine |
| Pediatrics | Pediatrics |
| Journal of the American Medical Informatics Association (JAMIA) | Medicine |
Related journal results evaluation of the content-similarity-based approach using different metrics_
| Number of papers published in 12 months | Number of Journals | CC CC: Pearson correlation coefficients; KTCC: Kendall’s tau correlation coefficients; NDCG: Normalized discounted cumulative gain. NDCG@5: NDCG with truncated position 5; NDCG@20: NDCG with truncated position 20. | KTCC CC: Pearson correlation coefficients; KTCC: Kendall’s tau correlation coefficients; NDCG: Normalized discounted cumulative gain. NDCG@5: NDCG with truncated position 5; NDCG@20: NDCG with truncated position 20. | F1 | NDCG@5 CC: Pearson correlation coefficients; KTCC: Kendall’s tau correlation coefficients; NDCG: Normalized discounted cumulative gain. NDCG@5: NDCG with truncated position 5; NDCG@20: NDCG with truncated position 20. | NDCG@20 CC: Pearson correlation coefficients; KTCC: Kendall’s tau correlation coefficients; NDCG: Normalized discounted cumulative gain. NDCG@5: NDCG with truncated position 5; NDCG@20: NDCG with truncated position 20. |
|---|---|---|---|---|---|---|
| >50 | 3,265 | 0.2617 | 0.0647 | 0.52 | 0.7235 | 0.6654 |
| >100 | 2,161 | 0.3271 | 0.1185 | 0.55 | 0.7583 | 0.7015 |
| >200 | 1,063 | 0.4153 | 0.1767 | 0.59 | 0.7931 | 0.7403 |
| >300 | 599 | 0.4646 | 0.1957 | 0.60 | 0.8003 | 0.7508 |
| >500 | 233 | 0.4591 | 0.1458 | 0.58 | 0.7570 | 0.7161 |
Top 20 journals related to Genetics identified by the citation-based, usage-based and content-similarity-based method, respectively_
| Citation-based | Usage-based | Content-similarity-based |
|---|---|---|
| PNAS | PNAS | PLOS Genetics |
| Cell | JBC | PNAS |
| Nature | Nature | MCB |
| MCB | Science | PLOS One |
| Science | MCB | Genome Research |
| TAG | Cell | Nature |
| Evolution | Development | Eukaryotic Cell |
| EMBO | Journal | Genes & Development Evolution |
| Genes & Development | NAR | MBoC |
| NAR | EMBO Journal | Molecular Ecology |
| JBC | Current Biology: CB | JBC |
| MBE | MBoC | Science |
| Journal of Bacteriology | Developmental Biology | Developmental Biology |
| MGG | MBE | Cell |
| Heredity | Nature Genetics | Current Biology: CB |
| Development | JCB | Genes & Development |
| Genetical Research | Journal of Bacteriology | Development |
| Genome | CCM | MBE |
| JMC | PLOS Genetics | Heredity |
| JCB | AJHG | AJHG |